Abstract
A mass spectrometric (MS)-based strategy for antigen (Ag) identification and characterization of globally produced monoclonal antibodies (mAbs) is described. Mice were immunized with a mixture of native glycoproteins, isolated from the pooled plasma of patients with non-small cell lung cancer (NSCLC), to generate a library of IgG-secreting hybridomas. Prior to immunization, the pooled NSCLC plasma was subjected to 3-sequential steps of affinity fractionation, including high abundant plasma protein depletion, glycoprotein enrichment and polyclonal antibody affinity chromatography normalization. In this paper, in order to demonstrate the high quality of the globally produced mAbs, we selected 3 mAbs of high differentiating power against a matched, pooled normal plasma sample. After production of large quantities of the mAbs from ascites fluids, Ag identification was achieved by immunoaffinity purification, SDS-PAGE, Western blotting and MS analysis of in-gel digest products. One antigen was found to be complement factor H, and the other two were mapped to different subunits of haptoglobin (Hpt). The 2 Hpt mAbs were characterized in detail in order to assess the quality of the mAbs produced by the global strategy. The affinity of one of the mAbs to the Hpt native tetramer form was found to have a KD of roughly 10−9 M and to be 2 orders of magnitude lower than the reduced form, demonstrating the power of the mAb proteomics technology in generating mAbs to the natural form of the proteins in blood. The binding of this mAb to the β-chain of haptoglobin was also dependent on glycosylation on this chain. The characterization of mAbs in this work reveals that the global mAb proteomics process can generate high-quality lung cancer specific mAbs capable of recognizing proteins in their native state.
Keywords: monoclonal antibody, mAb proteomics, antibody characterization, glycoprotein, haptoglobin, conformational epitope, lung cancer
Introduction
The search for clinical protein biomarkers in body fluids such as plasma continues to be an area of active research. There have been significant efforts to develop various platforms using liquid chromatography-mass spectrometry (LC-MS) -based shotgun proteomics to discover markers associated with disease.1-4 The dynamic range of plasma proteins, the heterogeneity due to post-translational modifications, and the overall sample complexity continue to be major challenges for shotgun proteomics-based platforms. While various strategies have been advanced, there is general agreement that the results to date have not realized the potential of the LC-MS approach, and validation of candidate markers for clinical application remains a major hurdle.3
One strategy for biomarker validation is to raise antibodies (Abs) to the discovered candidate markers for immunoassay analysis, e.g. ELISA. The ELISA method can be highly sensitive, can be a high throughput method, and is widely accepted as the gold standard in clinical medicine and biological research.5 On the other hand, ELISA development is time consuming, and the generation of Abs with high affinity and specificity cannot be certain. Recently, the use of well characterized monospecific polyclonal Abs as capture reagents has been proposed as a technology for clinical proteomics and biomarker validation6 and several initiatives are underway to generate Ab libraries.7,8 These technologies include the production of scFv antibody subunits using phage display7 and the generation of polyclonal Abs (pAbs) against recombinant peptides.9 In addition, the use of monoclonal Ab (mAb) libraries generated against mixtures of immunogens from tissue organelles10 or plasma11 has been suggested as an alternative (“reverse proteomic”) approach for the discovery of biomarkers.
Another global approach, namely mAb proteomics, has recently been introduced using a platform for generating large libraries of mAbs in a high throughput manner.12 Mice are immunized with complex biological mixtures of proteins, e.g. from enriched plasma. A large hybridoma library is generated, and the mAbs are subjected to high throughput ELISA screening to identify disease specific mAbs. These mAbs could potentially be directly used in antibody arrays to screen large sample populations. However, the characteristics of these globally generated mAbs must be evaluated, including the identification of their specific antigens. It is the purpose of this paper to first identify the antigens associated with specific mAbs, and then further characterize the quality of the mAbs produced to provide insight into the effectiveness of the global mAb production approach. We have established a workflow for the antigen identification procedure using a mass spectrometric-based method combined with a simple immunoassay, and then demonstrate that the mAbs generated by this approach are elicited to the native form of the protein as it exists in plasma.
Experimental
A Materials
Sodium cyanoborohydride, dimethyl pimelimidate•2HCl (DMP), EZ-link™ plus activated peroxidase were purchased from Thermo Fisher Scientific (San Jose, CA). POROS® Affinity Protein G beads and horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody were from Life Technologies (Carlsbad, CA). Sequencing grade trypsin was obtained from Promega (Madison, WI). Chemiluminescence HRP substrate was obtained from GE Healthcare (Buckinghamshire, UK). Colorimetric HRP substrate, tetramethylbenzidine (TMB), was from BioFX (Owings Mills, MD). All other reagents were obtained from Sigma-Aldrich (St. Louis, MO).
B Plasma sample collection and immunogen preparation
Plasma from 20 patients diagnosed with non-small cell lung cancer (NSCLC) and 20 healthy control patients, matched by age, gender and race, were obtained from Proteogenex (Culver City, CA). According to the histological diagnosis, 80% of the cancers were squamous cell carcinoma, 15% were adenocarcinoma, and 5% were undifferentiated NSCLC. All of the patients except one were free of distant metastasis, with primary tumors larger than 2 cm.
Equal volumes of plasma from each patient or matched control were pooled. The pooled plasma was subjected to 3-sequential steps of affinity fractionation. First, the concentration of the twelve most abundant plasma proteins was reduced using the Seppro LC10 IgY depletion immunoaffinity column (Sigma-Aldrich, St. Louis, MO). Then, the glycoproteins in the flow-through from the depletion column were enriched using a multi-lectin affinity column (M-LAC).13 Finally, the eluate from M-LAC column was subjected to a polyclonal affinity column for normalization.14 The normalized M-LAC material of native glycoproteins from the NSCLC plasma was used for immunization to produce the IgG-secreting hybridoma library.
C Hybridoma screening
Screening of IgG containing hybridoma supernatants was performed in 384 well, high-binding plates (Corning Inc., Lowell, MA), coated for 2h at room temperature (RT) with 20 μg/mL goat anti-mouse polyclonal antibody (13 μL/well, Southern Biotechnology Associates, Inc., Birmingham, AL) in carbonate-bicarbonate coating buffer at pH 9.6. The plates were blocked with 40 μL of 0.5% BSA in PBS at 4 °C overnight. Each IgG (from 13 μL non-diluted hybridoma supernatant) was added to four adjacent wells for independent readings. The plates contained eight positive (15 ng mouse IgG anti-human albumin) and eight negative (13 μL complete growth medium) controls. Following an overnight incubation at 4 °C and plate washing, 7 μL of biotinylated depleted plasma (10 μg/mL) in PBS with 0.05% (v/v) Tween 20 and 1% low IgG fetal bovine serum were added to all wells. Labeling of the proteins by biotin was performed on depleted plasma using a commercially available biotinylation kit (EZ-Link Sulfo-NHS-LC-Biotin kit, Thermo-Fisher, Rockford, IL). Seven μL of biotinylated human serum albumin (5 μg/mL) from Sigma was added to the wells of the positive controls. After 90 min incubation at RT, the unbound proteins were removed by washing the plates, and the signals were developed by addition of streptavidin-labeled-peroxidase and OPD peroxidase substrate (Vectastain Elite ABC Peroxidase kit, Vector Laboratories, Burlingame, CA.). The kinetic measurement of the reaction was performed with a microplate reader SpectraMax (Molecular Device, MDS, Toronto, Canada) at 450 nm for 4 min (37 °C). The Vmax of the chromegenic reactions (calculated from the linear part of the kinetic curve) were normalized across plates using the positive (PC) and negative controls (NC) according to the following expression: VmaxN sample = (Vmax sample −Vmax NC)/ (Vmax PC−Vmax NC).
D Immunoaffinity purification
A small subset of the IgG-secreting hybridoma cell lines that discriminated between lung cancer and matched controls was cloned according to standard protocols.15 Three mAbs (mAb #1, #2 and #3) were produced in large quantities from ascites fluids and purified by means of HiTrap Protein G (GE Healthcare) and Protein L (Thermo Fisher Scientific) affinity chromatography. The purified mAbs were crosslinked to POROS® Affinity Protein G beads using DMP and packed into PEEK columns (2 × 20 mm, 63 μL), respectively. For affinity purification, a Shimadzu HPLC system (LC-10AD, Columbia, MD) was used. Plasma was mixed with an equal volume of binding buffer (100 mM phosphate, 150 mM NaCl, pH 7.5), and 50 μL of sample was injected into the HPLC system. The unbound proteins were washed away with 5 column volumes of binding buffer, and then the retained proteins were eluted with 100 mM glycine (pH 2.5). The eluted proteins were collected directly into an Amicon spin column (MWCO: 5 kDa, Ultra-15, Bedford, MA) and buffer exchanged to PBS by means of centrifugation.
E SDS-PAGE and Western blotting
Using a NuPAGE® Novex® 4 - 12% Bis-Tris mini-gel system (Life Technologies), SDS-PAGE was conducted under either reduced or non-reduced conditions. For reduced SDS-PAGE, the affinity purified antigen was mixed with lithium dodecyl sulfate (LDS) sample buffer and reducing agent (dithiothreitol (DTT)) and then incubated at 70 °C for 10 mins before loading on to the gel. For non-reduced SDS-PAGE, the sample was mixed with LDS sample buffer without addition of DTT. Gels were stained with Coomassie blue for protein detection.
For Western blotting, the proteins on the SDS-PAGE gel were electroblotted onto a nitrocellulose membrane using XCell II Blotting Module (Life Technologies). The membrane was treated with blocking solution (0.5% BSA in PBS) for 1 hour with gentle shaking, followed by incubation with 1/2000 dilution of the mAb (stock concentration 1 mg/mL) in blocking solution for 1 hour. The membrane was quickly washed with 18 MΩ water, followed by 3 consecutive washes with TBS/ 0.05% v/v Tween (TBST) (10 min × 20 mL/each). The HRP-conjugated goat anti-mouse secondary antibody was 1/40,000 diluted in blocking solution, and the membrane was incubated for 1 hour, followed by washing as above. Chemiluminescence HRP substrate was incubated with the membrane for 2 min before detection using FluorChem™ Imaging System (Alfa Innotech, San Leandro, CA).
F In-gel and in-solution tryptic digestion
For in-gel digestion, the protein bands of interest were excised and minced into small pieces (~ 0.5 mm2). Coomassie dye was removed by 2-3 cycles wash with acetonitrile and rehydration with 100 mM ammonium bicarbonate. After destaining, the proteins were reduced by incubation with 10 mM DTT for 30 min at 56 °C and then alkylated with 55 mM iodoacetamide (IAA) for 60 min at room temperature in the dark. Trypsin was added to the gel pieces and incubated for 40 mins on ice. The unabsorbed trypsin solution was replaced with 50 mM ammonium bicarbonate, and sufficient volume was added to cover the gel pieces. Samples were incubated overnight at 37 °C, and then the supernatant was removed and stored. Gel pieces were further extracted with 5% formic acid at 37 °C for 15 min with constant shaking. The extracted solution was combined with the stored supernatant and completely dried using a Speed Vac. Peptides were redissolved in 10 μL 0.1% formic acid for LC-ESI-MS or MALDI-TOF-MS analysis.
For in-solution digestion, the protein solution (IP pull-down mixture, M-LAC material or normalized MLAC material) was denatured by 6M guanidine·HCl, reduced by 10 mM DTT (30 min at 56 °C) and alkylated by 50mM IAA (60 min at room temperature in the dark). The reduced and alkylated protein solution was buffer exchanged to 50 mM ammonium bicarbonate using Microcon ultracentrifuge device (5 kDa MWCO, Millipore, Billerica, MA) for 5 cycles (10 000× g for 5 min per cycle). Trypsin was added into the protein solution at ~ 1:25 ratio (enzyme to protein) and incubated for 6 hrs. The enzymatic reaction was stopped by 5% formic acid and the digest was stored at −80 °C until analysis.
G LC-ESI-MS and MALDI-TOF-MS analysis
For LC-ESI-MS, an LTQ-FT mass spectrometer (Thermo Fisher Scientific) interfaced with an Ultimate 2000 nanoLC (Dionex, Sunnyvale, CA) was used. A 75 μm i.d. capillary column (New Objective, Woburn, MA) was packed in-house with Magic C18 (3 μm, 200 Å pore size) stationary phase (Michrom Bioresources, Auburn, CA). Peptides were eluted at a flow rate of 200 nL/min, and MS/MS spectra were obtained in a data-dependent mode in which the 8 most intense peaks in each MS scan were chosen for fragmentation. An m/z width of +/− 1 Da was employed to isolate the peptide precursor ions, and a 35% normalized collision energy was used to fragment the isolated peptides. Dynamic exclusion was utilized with exclusion duration of 30 s and no repeat counts. Protein identification was achieved by searching the Swiss-Prot human protein database using SEQUEST.
MALDI-TOF mass spectra were acquired using an Applied Biosystems 4700 TOF/TOF Proteomics Analyzer equipped with delayed extraction and a 200-Hz repetition rate UV laser (355 nm). The instrument was externally calibrated using angiotensin I. The matrix, α-cyano-4-hydroxycinnamic acid (CHCA, Mass PREP, Waters, Milford, MA), was prepared in acetonitrile/water, containing 0.1% trifluoroacetic acid (TFA) (50:50, v/v), at a concentration of 7 mg/mL. 0.5 μL of the in-gel digest were spotted onto the MALDI plate with 0.4 μL matrix using a thin-layer spotting method. Protein identification was accomplished by searching the Swiss-Prot human protein database using MASCOT in the peptide mass fingerprinting (PMF) mode.
H Deglycosylation and glycan-specific staining
Enzymatic deglycosylation using PNGase F was achieved in either native or reduced conditions. For deglycosylation of the native protein, affinity purified antigen was mixed with 250 mM phosphate buffer (pH 7.5) at a 1:4 ratio, followed by the addition of PNGase F (1 Sigma unit enzyme for 20 μg Hpt) and incubation at 37 °C for up to 4 days. Two additional aliquots of PNGase F were transferred into the reaction mixture at the end of the 2nd and 3rd days, respectively. For deglycosylation under reduced conditions, the sample was mixed with reaction buffer (250 mM phosphate buffer, pH 7.5) and denaturation buffer (1% SDS and 1% mercaptoethanol), and then heated at 100 °C for 5 mins. After cooling to room temperature, PNGase F was added (1 Sigma unit enzyme for 20 μg Hpt), and the mixture was allowed to incubate at 37 °C for 3 - 4 hours with constant shaking.
Glycan-specific staining was accomplished using periodic acid and a Schiff's base reagent.16 Briefly, the gel was first fixed overnight with 40% ethanol and 7% acetic acid in order to minimize protein diffusion. Glycans attached to the glycoprotein were oxidized by 1% periodic acid and 3% acetic acid for 1 hour to generate aldehyde groups. The gel was then washed thoroughly with deionized water to remove periodic acid. After washing, the gel was incubated with Schiff's base reagent for 1 hour in the dark. A solution of 0.6% potassium metabisulfite and 3% acetic acid was used to wash the gel in order to prevent color fading.
I Surface plasmon resonance (SPR) analysis
Surface plasmon resonance was performed on a T100 instrument (Biacore, Uppsala, Sweden). The sensor chip (type CM5, series S) surface was first activated with a 1:1 mixture of 0.2 M N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide (EDC) and 0.05 M N-hydroxysuccinimide (NHS) in water, followed by a goat anti-mouse antibody solution (30 μg/mL in 10mM sodium acetate, pH 5.0). The goat anti-mouse antibody (mouse antibody capture kit, Biacore) was immobilized on the sensor chip surface through amine coupling, and the remaining unreacted NHS binding sites were blocked with 1 M ethanolamine-HCl, pH 8.5. For SPR analysis, mAb was first passed through the chip channel, and, after washing with running buffer, the serially diluted sample was introduced and the interaction recorded in real-time. Regeneration of the sensor surface between analysis cycles was achieved using 10 mM glycine-HCl, pH 1.7. The running buffer was 10 mM HEPES, 150 mM NaCl, 3.0 mM CaCl2, 1.0 mM EGTA, and 0.005% Tween-20, pH 7.4. All binding data were analyzed using the Biomolecular Interaction Analysis evaluation program version 3.1 (Biacore).
J Sandwich ELISA
mAb #1 was conjugated to HRP as follows: a total of 100 μg of EZ-link™ plus activated peroxidase was reconstituted in 10 μL ultrapure water and added to the 100 μL antibody solution (1 mg/mL mAb #1 in PBS, pH 7.8). Immediately, 1.1 μL of freshly-prepared sodium cyanoborohydride (0.5 M in PBS) was added to the antibody solution, and the solution was then incubated for 3 hours at R.T. (final concentration of sodium cyanoborohydride, 5 mM). Four μL of 3 M ethanolamine was added to the antibody solution to quench the reaction. The reaction solution was buffer exchanged to PBS and incubated overnight with Sepharose Protein G beads (Thermo Fisher Scientific) at 4 °C. The HRP-mAb #1 conjugate was captured and eluted from the Protein G column using 0.1 M glycine, pH 2.5, and the eluate was immediately neutralized with 1 M Tris (1/10 volume), followed by buffer exchange to PBS. The activity of the HRP-mAb #1 conjugate was confirmed by its ability to turn the TMB substrate from colorless to blue. For the sandwich ELISA, mAb #2 (20 μg/mL in PBS) was coated onto a 96-well plate. Serially diluted standard Hpt solution (0.005 - 10 μg/mL) was added into each well, followed by incubation with HRP-mAb #1 conjugate for standard curve generation. A negative control (blank) was used to monitor the non-specific signal. For Hpt concentration determination in crude plasma, the plasma was diluted 600 times with PBS. For Hpt concentration determination in M-LAC material, the resulting M-LAC material was directly used with no further treatment.
Results and Discussion
The global production of mAb libraries to complex mixtures of proteins derived from diseased samples such as cancer can potentially be developed as diagnostic immunoassay kits after the use of the mAbs for the discovery and ultimate validation of biomarkers. However, the antigens should first be identified, and the antibodies characterized. The focus of this work was to develop a platform for antigen identification and then to characterize the antibodies to assess the effectiveness of the mAb proteomics approach. The generation of hybridoma libraries and subsequent screening for lung cancer specific hybridomas will be detailed in a separate paper. Briefly, mice were immunized with NSCLC patient plasma that was depleted of major proteins, glycoprotein enriched and further normalized. This non-targeted, global immunization strategy generated a large library (1055) of IgG-secreting hybridomas. The hybridomas were subjected to high throughput ELISA screening, and a large number of hybridomas (184) were found to discriminate between pooled lung cancer and matched normal samples at an abundance difference level of 1.5. The initial step of screening the hybridomas with pooled plasma was necessary because of the limited amount of supernatant collected at the initial stages of the hybridoma growth. Subsequent screening of individual plasma samples with supernatants and/or the purified IgG from the cloned cell lines identified 13 mAbs with strong differentiating powers (p-values calculated from Mann-Whitney U test < 0.01). Three of the cloned mAbs had an estimated difference of 3.5, 2.5 and 3.0 fold in abundance for mAb #1, #2 and #3, respectively, between lung cancer and matched controls. These 3 mAbs were then produced in large quantities from ascites fluids for antigen identification. Antigen identification was achieved using a workflow consisting of immunoaffinity purification (IP pull-down), SDS-PAGE, Western blotting and MS-based analysis, as described below.
1. Antigen identification
Immunoprecipitation followed by MS analysis is typically used for antigen identification,10-11 but during affinity purification, not only the antigen but also its natural interacting partners and non-specific “sticky” proteins can be co-precipitated with the antigen. As a result, MS analysis will not directly yield a single protein identification, but rather a list of proteins. For example, when mAb #1 was used to IP pull-down the antigen from plasma, after trypsin digestion and LC-ESI-MS analysis, more than 30 proteins were identified. The top 9 proteins (Table 1) were all identified with more than 10 peptide digest fragments. Such a result makes it challenging to identify the antigen unambiguously and requires additional steps. In one case,10 a cDNA expression library was used to screen generated Abs, and the results from the screening were correlated with MS data to determine the antigen identity.
Table 1.
Proteins Identified from the IP Pull-Down Mixture Using mAb #1*
| Protein Name | Peptide Counts | |
|---|---|---|
| 1 | Haptoglobin | 103 |
| 2 | Serum albumin | 33 |
| 3 | C4b-binding protein α chain | 29 |
| 4 | Apolipoprotein A-I | 26 |
| 5 | Hemoglobin subunit β | 20 |
| 6 | Hemoglobin subunit α | 13 |
| 7 | Ig alpha-1 chain C region | 11 |
| 8 | Ig kappa chain C region | 10 |
| 9 | Ig gamma-3 chain C region | 10 |
The criteria used are as following: Xcorr >=1.9 when charge is +1, >= 2.2 when charge is +2, >= 3.8 when charge is +3; ΔCn>=0.1 and Peptide Prophet>=0.95.
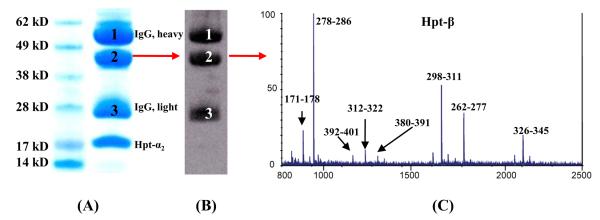
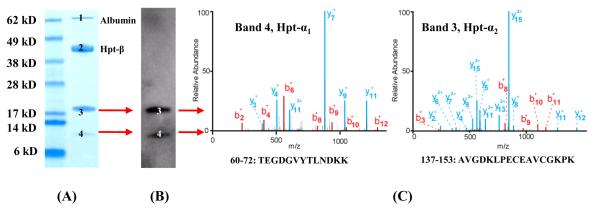
Here we used Western blotting to identify the protein band from the IP pull-down, followed by MS analysis for the antigen identification. As shown in Figure 1, the antigen protein was first isolated from plasma using an immunoaffinity column with the specific mAb; the eluate was then separated by SDS-PAGE. Two separate gels were run: one was used for Coomassie blue staining and the other for Western blotting. The Western blotting membrane was probed with the same mAb used in the immunoaffinity purification step, and the protein band detected by the antibody was targeted for MS analysis. The theoretical molecular weight of the identified protein was also compared with that of the SDS-PAGE / Western blotting results. mAb #1 was found to target the Hpt-β chain. The identification of the antigen using MALDI-TOF-MS is shown in Figure 2, with a total of 11 tryptic peptides (56% sequence coverage of the β-chain) identified with a mass accuracy of better than 20 ppm. The major peaks from Hpt-β are annotated in Figure 2-C. The mAb #2 was determined to target Hpt-α (Figure 3), with both the α1 and α2 chains of Hpt17 being recognized by this mAb. The identification was accomplished using LC-ESI-MS. The sequence coverage for Hpt-α1 and Hpt-α2 are 40% and 52%, respectively. Two representative MS/MS spectra resulting from Hpt-α1 (Figure 3-A, band 4,) and Hpt-α2 (Figure 3-A, band 3) are shown in Figure 3-C. The peptide fragments (b- and y- ion series, Figure 3-C) unambiguously determined the identity of the antigen. In plasma, native Hpt circulates as an (αβ)2 tetramer with N-linked glycosylation sites on its β-chain.17 The α-chain is polymorphic, and Hpt-α2 is the product of gene duplication with approximately twice the length of Hpt-α1.18,19
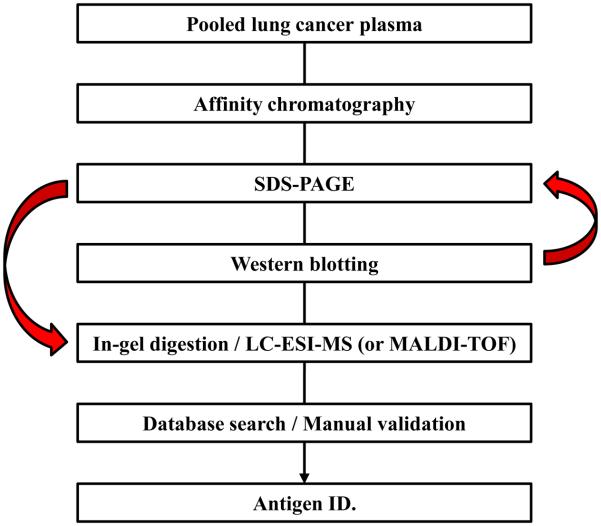
Figure 1. Antigen Identification Workflow.
The affinity purified antigen mixture was separated by SDS-PAGE, and then probed with a specific mAb to locate the binding antigen. The MW region, which was reactive in Western blotting, was excised from the gel, digested with trypsin and sequenced by mass spectrometry.
Figure 2. Antigen Identification for mAb #1.
Reduced SDS-PAGE of the mAb #1 affinity purified mixture from pooled lung cancer patient plasma (15 μg) and stained with Coomassie blue (A). Bands 1 and 3 are the heavy (~ 50 kDa) and light chains (~ 25 kDa) of IgG, respectively. The protein (band 2) migrating at ~ 44 kDa was recognized by the mAb #1 (B) and was subsequently identified as Hpt-β chain by MALDI-TOF-MS (C).
Figure 3. Antigen Identification for mAb #2.
Reduced SDS-PAGE of an affinity purified mixture (5 μg) from IgG- and albumin-depleted pooled lung cancer plasma, stained with Coomassie blue (A). Bands 1 and 2 are albumin and Hpt-β chain, respectively. Bands 3 (~18 kDa) and 4 (~10 kDa) were both recognized by mAb #2 (B) and were subsequently identified as the Hpt-α2 and Hpt-α1 chain by LC-ESI-MS (C).
Haptoglobin (either Hpt-α,20 Hpt-β21-23 or whole Hpt protein24) has previously been suggested to be candidate marker for various cancers, including lung cancer.24 However, in our study, we expected that the majority of Hpt from the lung cancer plasma would be removed following immunodepletion with the IgY-antibodies. We performed a shotgun proteomic analysis (see Experimental Section) on the M-LAC and normalized M-LAC fractions to estimate the amount of Hpt in the two samples. It is interesting to note that Hpt was identified in both samples with total peptide counts of 14 and 3 for the M-LAC and normalized M-LAC fractions, respectively. This result indicates the presence of residual amounts of Hpt in the samples used for immunization (according to the manufacturer, the removal efficiency of Hpt from the depletion column is not complete but is > 95%). To accurately determine the levels of Hpt, we measured the Hpt concentration in the M-LAC sample (the protein mixture after M-LAC glycoprotein enrichment) using a sandwich ELISA assay developed with mAbs #1 and #2 (see below). The concentration of Hpt in M-LAC sample was determined to be ~120 ng/mL. Unfortunately, there was not sufficient amount of normalized M-LAC sample for direct Hpt concentration measurement by ELISA. However, it can be estimated that the Hpt injected into the mouse was in the low to medium ng/mL level. This estimation suggests that this global method of immunization has the potential of eliciting an immune response to low abundance proteins, such as those originating from tumor cells but secreted into the bloodstream, if proper sample pretreatment is achieved. In this study, we have only focused on 3 of the discriminating mAbs; however, at least 184 hybridomas have been shown to discriminate between lung cancer and matched controls. Future antigen identification of this set of mAbs could reveal potentially novel markers for NSCLC.
Turning to mAb #3, the antigen was identified in a similar manner as above and found to be complement factor H (CFH, Supplementary Material, Figure S1). The characterization of mAb #3 was not pursued in this study.
2. Western blotting
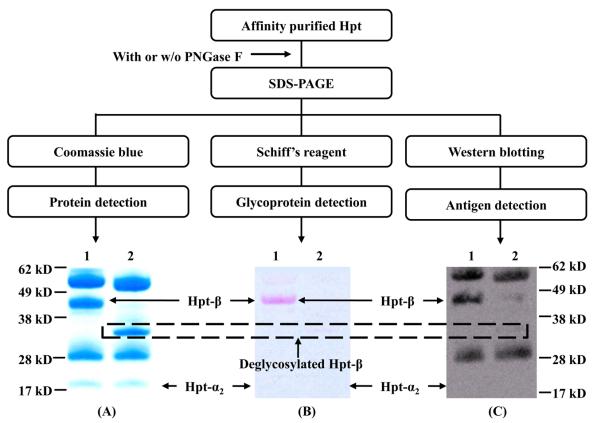
To test whether the affinity of anti-Hpt-β was influenced by Hpt glycosylation, the binding of mAb #1 was examined for both the glycosylated and deglycosylated form of Hpt. PNGase F was used to remove the N-linked sugars from Hpt, and the deglycosylated protein was subsequently examined by reduced SDS-PAGE and Western blotting. The protein was visualized by Western blotting, and the protein band corresponding to the signal observed on the blot was excised from the gel, digested and analyzed by mass spectrometry. The identified Hpt-β chain has a MW ~ 44 kDa (Figure 2-A, band 2), which is 13 kDa larger than its theoretical MW (31 kDa), due to the glycosylation attached to the chain. It is well known that glycans attached to an antigen could affect Ab-Ag interaction,25,26 either directly through hydrogen bonding and/or electrostatic interaction, or indirectly by glycosylation-induced conformational changes of the protein. As shown in Figure 4, the Hpt-β chain is seen to migrate with a MW of 44 kDa (Figure 4-A, lane 1); after PNGase F treatment, Hpt-β shifted to an apparent MW of 31 kDa (Figure 4-A, lane 2). Glycan-specific staining confirmed that the shift in MW was due to the removal of the glycans (Figure 4-B). A separate Hpt preparation, which underwent all the deglycosylation steps except for the addition of PNGase F, was run in parallel with the deglycosylated Hpt as a positive control (Figure 4-C, lane 1). Western blotting indicated that N-linked glycosylation on Hpt-β is essential for the Ab-Ag interaction (Figure 4-C, lane 2). In a separate experiment, Hpt was subjected to PNGase F treatment under native conditions, followed by non-reduced SDS-PAGE and Western blotting (Supplementary Material, Figure S2). As expected, the complete removal of all glycans from the native Hpt was not possible, even after a long incubation time (4 days) and repetitive additions of PNGase F. Nonetheless, the signal for Western blotting of the Ab-Ag interaction was weaker for the partially deglycosylated protein, further suggesting that the glycan structures on Hpt-β chain are necessary for the Ab-Ag interaction.
Figure 4. N-linked Glycosylation is Essential for the mAb-Ag Interaction.
Glycosylated and deglycosylated haptoglobin were separated by SDS-PAGE and stained with either Coomassie blue for protein detection (A), Schiff's base reagent to determine the deglycosylation efficiency (B) or transferred to the nitrocellulose membrane for Western blotting (C, probed with anti-Hpt-β mAb #1).
3. Surface plasmon resonance
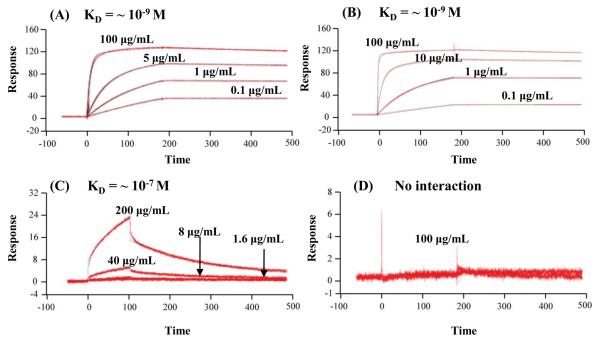
Surface plasmon resonance (SPR) was employed to measure the affinity constant (KD) of mAb #1 (anti-Hpt-β) against native Hpt purified from the pooled lung cancer and matched control plasma samples. Multiple rounds of affinity purification were conducted to obtain sufficiently pure Hpt (data not shown). Anti-Hpt-β was immobilized onto two flow channels of the same SPR sensor chip, one for the sample and the other to serve as a reference channel or control. A serially diluted affinity purified native Hpt sample was introduced, and the interaction was recorded in real time. A similar KD (10−9 mol) of anti-Hpt-β against native Hpt (glycosylated tetrameric form) was observed for both the lung cancer and matched control samples (Figures 5 A-B). To further characterize the reactivity of anti-Hpt-β, we measured the KD of anti-Hpt-β with a) reduced Hpt-β chain (glycosylated monomeric form, Figure 5-C) and b) reduced and deglycosylated Hpt-β chain (deglycosylated monomeric form, Figure 5-D). Due to the limited availability of lung cancer patient plasma samples, these SPR experiments were conducted using Hpt isolated from pooled, untreated normal plasma. The results indicated that mAb #1 binds with relatively tight affinity to the native Hpt tetramer, with a KD of ~ 10−9 M (Figure 5-B), versus 2 orders of magnitude lower for the reduced Hpt (KD of ~ 10−7 M, Figure 5-C). As expected, no interaction was observed for reduced and deglycosylated Hpt (Figure 5-D). The affinity constant of mAb #2 (anti-Hpt-α) against native Hpt was also measured by SPR and the KD was found to be in the 10−8 M range.
Figure 5. Surface Plasmon Resonance Analysis of Anti-Hpt-β with Hpt.
Panels A-D are sensorgrams of the interaction of anti-Hpt-β with (A) native Hpt (lung cancer); (B) native Hpt (matched control); (C) reduced Hpt-β chain (matched control); and (D) reduced and deglycosylated Hpt-β chain (matched control).
The favorable interaction of mAb #1 with the tetrameric form in comparison to the monomeric form suggests that anti-Hpt-β was generated to the protein that actually exists in plasma, i.e., the native form. This is an important result since mAbs are typically produced against synthetic peptides or recombinant protein fragments which generally are not the native form. No interaction between anti-Hpt-β and the reduced and deglycosylated Hpt was observed, further suggesting that the mAb proteomics process generates mAbs to the protein as it exists in plasma. Ignoring serum albumin, over 80% of the plasma proteome is glycosylated,27, 28 and many of these glycoproteins require their glycans to be functionally active. In summary, we have demonstrated that the mAb proteomics process can produce large numbers of monoclonal antibodies, some of which are of high affinity (KD in nmol range) that target the native structure and associated PTMs.
It is interesting to note that similar affinity constants of anti-Hpt-β were observed for both the lung cancer and matched control samples. Aberrant glycosylation changes on Hpt-β chain have been found for various cancers,22-24 including lung cancer.24 This result seems to suggest that, although the interaction of anti-Hpt-β mAb and the Hpt protein is affected by the presence of glycan structures, the high affinity of anti-Hpt-β may not be specific for cancer-induced glycosylation changes. Thus, the high differentiating power of this mAb for lung cancer vs. normal plasma seems to mainly result from the Hpt's abundance difference between the lung cancer and matched control sample.
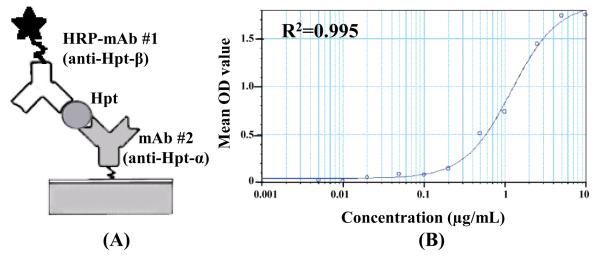
4. Sandwich ELISA of Hpt
The sandwich ELISA assay is taken to be the gold standard for measuring the concentration of proteins in crude and complex mixtures; however, the development of an ELISA assay requires highly specific antibody pairs targeting separate epitopes on the antigen, often a difficult goal to achieve. In this study, since mAb #1 and mAb #2 were found to target different subunits on the same protein, the potential to develop an ELISA assay was examined. mAb #1 was labeled in-house with HRP and used as the detection antibody, as shown in Figure 6-A. Activated HRP was chosen to label the antibody, and the resulting Schiff base was reduced to form a stable secondary amine with sodium cyanoborohydride. A standard sandwich ELISA operation procedure was followed,5 and the calibration curve was generated using Hpt ranging from 0.005 - 10 μg/mL (Figure 6-B). The sandwich ELISA assay was used to measure a) the Hpt concentration in the M-LAC sample from pooled lung cancer patients plasma and b) the Hpt levels in crude plasma of a subset of individual lung cancer patients and controls. For the M-LAC sample, as noted earlier, the Hpt concentration was found to be ~ 120 ng/mL. For the individual samples, the Hpt concentration in the lung cancer patients was found to be ~ 3 mg/mL and in the normal samples to be 1 mg/mL (Table 2). This result confirmed the expected up-regulation of Hpt in the plasma of the lung cancer patients and served as an independent check of the cancer-specific mAb screening. The high number of mAbs generated by the mAb proteomics clearly increases the chances of producing mAb pairs suitable for the development of sandwich ELISA assays. This can be an important potential advantage of this method.
Figure 6. Sandwich ELISA Using mAb #1 and mAb #2.
(A) mAb #1 was labeled with HRP for detection and mAb #2 was used as the capture Ab. (B) The standard curve was generated using Hpt standard ranging from 0.005 - 10 μg/mL. The haptoglobin concentration in M-LAC material, lung cancer and matched control plasma sample were measured using this assay.
Table 2.
Hpt Concentration Determined from Individual Lung Cancer and Healthy Plasma
| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| Gender | M | M | M | M | M |
| Diagnosis | Healthy | Healthy | Lung cancer* | Lung cancer* | Lung cancer* |
| Smoking | No | 30 years, 1 pack/day | No | 45 years, 1.5 pack/day | 35 years, 1 pack/day |
| Hpt Conc. (mg/mL) |
0.82 | 0.76 | 3.4 | 2.86 | 3.2 |
All the lung cancer samples are squamous cell carcinoma without metastasis.
Conclusions
In this work, we used Western blotting and mass spectrometry to identify antigens from globally produced mAbs and characterized Ab-Ag interaction using Western blotting, SPR and sandwich ELISA. The Western blotting results indicated that the reactivity of mAb to the beta chain of Hpt was affected by the glycosylation on the chain. Surface plasmon resonance revealed that this mAb was generated to the native form of the protein in plasma with relatively tight binding (KD in the low nM range). Also, using a sandwich ELISA from two mAbs of Hpt and LC-MS based proteomics analysis, the Hpt concentration in the immunogen mix was determined to be in the low ng/mL range, demonstrating that the mAb proteomics used in this work can generate mAbs against low abundance plasma proteins, e.g. potentially proteins originating from tumor cells. The results of this study indicate that the mAb proteomics process is a promising approach for the high throughput generation of disease specific mAbs.
Supplementary Material
Acknowledgement
This work was supported by NIH grant CA126220 and in part GM15847. The authors thank Albert Tai, Tufts University, for his help on the SPR experiment. Contribution number 953 from the Barnett Institute.
Footnotes
Supporting information available
This material is available at http://pubs.acs.org.
References
- 1.Werner ZJ, Hanno L. How industry is approaching the search for new diagnostic markers and biomarkers. Mol. Cell. Proteomics. 2004;3:345–354. doi: 10.1074/mcp.M400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol. Cell. Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotech. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 4.Keshishian H, Addona T, Burgess M, Kuhn E, Carr S. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldsby R, Kindt T, Osborne B. Kuby immunology. 4th edition. W.H. Freeman publishing; 2002. pp. 148–150. chapter 6. [Google Scholar]
- 6.Uhlen M. Mapping the human proteome using antibodies. Mol. Cell. Proteomics. 2007;6:1455–1456. [PubMed] [Google Scholar]
- 7.Schofield D, Pope A, Clementel V, Buckell J, Chapple S, Clarke K, Conquer J, Crofts A, Crowther S, Dyson M, Flack G, Griffin G, Hooks Y, Howat W, Kolb-Kokocinski A, Kunze S, Martin C, Maslen G, Mitchell J, O'Sullivan M, Perera R, Roake W, Shadbolt S, Vincent K, Warford A, Wilson W, Xie J, Young J, McCafferty J. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254 1–18. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taussig M, Stoevesandt O, Borrebaeck C, Bradbury A, Cahill D, Cambillau C, Daruvar A, Dubel S, Eichler J, Frank R. ProteomeBinders: planning a european resource of affinity reagents for analysis of the human proteome. Nat. Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 9.Agaton C, Galli J, Guthenberg I, Janzon L, Hansson M, Asplund A, Brundell E, Lindberg S, Ruthberg I, Wester K, Wurtz D, Höög C, Lundeberg J, Ståhl S, Pontén F, Uhlén M. Affinity proteomics for systematic protein profiling of chromosome 21 gene products in human tissues. Mol. Cell. Proteomics. 2003;2:405–414. doi: 10.1074/mcp.M300022-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Gao J, Gao Y, Ju Y, Yang J, Wu Q, Zhang J, Du X, Wang Z, Song Y, Li H, Luo X, Ren F, Li J, Chen Y, Wang L, Xu H, Liu X, Wang J, Zhang Y, Cai Y, Cui Y, Qian X, He F, Li M, Sun Q. Proteomics-based generation and characterization of monoclonal antibodies against human liver mitochondrial proteins. Proteomics. 2006;6:427–437. doi: 10.1002/pmic.200500409. [DOI] [PubMed] [Google Scholar]
- 11.Ning Y, Wang Y, Li Y, Hong Y, Peng D, Liu Y, Wang J, Hao W, Tian X, Wu F, Dong W, Wang L, Wu Q, Liu X, Gao J, He F, Qian X, Sun Q, Li M. An alternative strategy for high throughput generation and characterization of monoclonal antibodies against human plasma proteins using fractionated native proteins as immunogens. Proteomics. 2006;6:438–448. doi: 10.1002/pmic.200500327. [DOI] [PubMed] [Google Scholar]
- 12.Csanky E, Olivova P, Rajnavolgyi E, Hempel W, Tardieu N, Katalin E, Jullien A, Malderez-Bloes C, Kuras M, Duval M, Nagy L, Scholtz B, Hancock W, Karger BL, Guttman A, Takacs L. Monoclonal antibody proteomics: discovery and prevalidation of chronic obstructive pulmonary disease biomarkers in a single step. Electrophoresis. 2007;28:4401–4406. doi: 10.1002/elps.200700256. [DOI] [PubMed] [Google Scholar]
- 13.Plavina T, Wakshull E, Hancock WS, Hincapie M. Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J. Proteome Res. 2007;6:662–671. doi: 10.1021/pr060413k. [DOI] [PubMed] [Google Scholar]
- 14.Takacs L, Guttman A, Kuras M. Normalization of complex analyte mixtures. 2007 PCT/IB2006/003161. [Google Scholar]
- 15.Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 16.Hardonk M, Duijn P. The Mechanism of the Schiff reaction as studied with histochemical model systems. J. Histochem. Cytochem. 1964;12:748–751. doi: 10.1177/12.10.748. [DOI] [PubMed] [Google Scholar]
- 17.Kurosky A, Barnett D, Lee T, Touchstone B, Hay R, Arnott M, Bowman B, Fitch W. Covalent structure of human haptoglobin: a serine protease homolog. PNAS. 1980;77:3388–3392. doi: 10.1073/pnas.77.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon G, Black J. Amino-acid sequence of alpha chains of human haptoglobins. Nature. 1968;218:736–741. doi: 10.1038/218736a0. [DOI] [PubMed] [Google Scholar]
- 19.Maeda N, Yang F, Barnett D, Bowman B, Smithies O. Duplication within the haptoglobin Hp2 gene. Nature. 1984;309:131–135. doi: 10.1038/309131a0. [DOI] [PubMed] [Google Scholar]
- 20.Ye B, Cramer D, Skates S, Gygi S, Pratomo V, Fu L, Horick N, Licklider L, Schorge J, Berkowitz R, Mok S. Haptoglobin-α subunit As potential serum biomarker in ovarian cancer: identification and characterization using proteomic profiling and mass spectrometry. Clin. Cancer Res. 2003;9:2904–2911. [PubMed] [Google Scholar]
- 21.Saito S, Murayama Y, Pan Y, Taima T, Fujimura T, Murayama K, Sadilek M, Egawa S, Ueno S, Ito A, Ishidoya S, Nakagawa H, Kato M, Satoh M, Endoh M, Arai Y. Haptoglobin-beta chain defined by monoclonal antibody RM2 as a novel serum marker for prostate cancer. Int. J. cancer. 2008;123:633–640. doi: 10.1002/ijc.23490. [DOI] [PubMed] [Google Scholar]
- 22.Okuyama N, Ide Y, Nakano M, Nakagawa T, Yamanaka K, Moriwaki K, Murata K, Ohigashi H, Yokoyama S, Eguchi H, Ishikawa O, Ito T, Kato M, Kasahara A, Kawano S, Gu J, Taniguchi N, Miyoshi E. Fucosylated haptoglobin is a novel marker for pancreatic cancer: A detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int. J. cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 23.Fujimura T, Shinohara Y, Tissot B, Pang P, Kurogochi M, Saito S, Arai Y, Sadilek M, Murayama K, Dell A, Nishimura S, Hakomori S. Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int. J. cancer. 2008;122:39–49. doi: 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- 24.Hoagland L, Campa M, Gottlin E, Herndon J, Patz E. Haptoglobin and posttranslational glycan-modified derivatives as serum biomarkers for the diagnosis of nonsmall cell lung cancer. Cancer. 2007;110:2260–2268. doi: 10.1002/cncr.23049. [DOI] [PubMed] [Google Scholar]
- 25.Katz Y, Guterman M, Lahat E. Regulation of synthesis of complement proteins in HEp2 cells. Clin. Immunol. Immunopathol. 1993;67:117–123. doi: 10.1006/clin.1993.1053. [DOI] [PubMed] [Google Scholar]
- 26.Milland J, Yuriev E, Xing P, McKenzie I, Ramsland P, Sandrin M. Carbohydrate residues downstream of the terminal Galalpha(1,3)Gal epitope modulate the specificity of xenoreactive antibodies. Immunol. Cell Biol. 2007;85:623–632. doi: 10.1038/sj.icb.7100111. [DOI] [PubMed] [Google Scholar]
- 27.Van P, Rudd P, Dwek R, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 28.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.