Abstract
Four cytokine receptor genes are located on Chr21q22.11, encoding the α and β subunits of the interferon alpha receptor (IFNAR1 and IFNAR2), the β subunit of the interleukin 10 receptor (IL10RB), and the second subunit of the interferon gamma receptor (IFNGR2). We previously reported that two variants in IFNAR1 were associated with susceptibility to malaria in Gambians. We now present an extensive fine-scale mapping of the associated region utilizing 45 additional genetic markers obtained from public databases and by sequencing a 44kb region in and around the IFNAR1 gene in 24 Gambian children (12 cases/ 12 controls). Within the IFNAR1 gene a newly studied C→G single nucleotide polymorphism (IFNAR1 272354c-g) at position −576 relative to the transcription start was found to be more strongly associated with susceptibility to severe malaria. Association was observed in three populations: in Gambian (P=0.002), Kenyan (P=0.022) and Vietnamese (P=0.005) case control studies. When all three studies were combined, using the Mantel-Haenszel test, the presence of IFNAR1 −576G was associated with a substantially elevated risk of severe malaria (N=2444, OR=1.38, 95%CI: 1.17-1.64; P=1.7×10−4). This study builds on previous work to further highlight the importance of the type I interferon pathway in malaria susceptibility and illustrates the utility of typing SNPs within regions of high linkage disequilibrium in multiple populations to confirm initial positive associations.
Keywords: gene, association, IFNAR1, severe malaria
Introduction
Malaria is a predominantly tropical disease and a global health problem in approximately one hundred countries. It affects two and a half billion people worldwide (approximately 40 percent of the world’s population) 1. The importance of host genetics in the study of the pathogenesis of malaria is underlined by the discovery of the protective effect of the sickle cell trait 2, the A- allele of Glucose-6-phosphate dehydrogenase (G6PD) 3, α-thalassemia 4, and more recently the haemoglobin E trait in peoples of South-East Asia 5, 6. Apart from molecules involved in erythrocyte physiology, recent work has revealed associations linking cytokines and their receptors to malaria pathogenesis 7. Examples include tumour necrosis factor (TNF)-α 8, 9 whereby a variant in the promoter confers susceptibility to severe malaria, and a subunit of the interferon gamma receptor (IFNGR1) in which heterozygosity at the locus may be associated with protection against severe malaria 10. Recently, genome-wide scans in the Plasmodium falciparum genome 11, 12 have identified putative targets for vaccination 13, further underlining the role of genetics in elucidating the pathogenesis and future preventive treatment of malaria.
Over the years, multiple studies have confirmed the crucial role of cytokines in the pathogenesis and clinical manifestation of malaria. High baseline levels of interleukin (IL)-10 has been found to associate with more severe clinical manifestations 14 and was convincingly replicated 15. Mouse models have suggested that interferon (IFN)-α may inhibit P. yoelii blood stage malaria by inhibiting reticulocyte production 16. It has been shown that IFNAR deficient mice lacked IFN-γ production in the presence of normal CD8+ T cell expansion 17 and co-immunoprecipitation studies have also suggested an interaction between IFNAR1 and IFNGR2 18, thus strengthening the link between IFN-α and IFN-γ signalling. The very well described IFN- γ has been shown to be a key cytokine in elucidating a strong TH1-biassed immune response, which is crucial in the clearance of intra-cellular pathogens, especially tuberculosis and invasive bacterial infections. More recently, IFN-γ has been demonstrated to play an important role in both parasite clearance and protective immunity to malaria 19, 20.
In recent years, there has been increasing interest in the relationship between Toll-like receptors (TLRs) (and members of their downstream signalling pathway) and the pathogenesis of different infectious diseases. Being an integral component of the innate immune response, it has recently been hypothesised that the TLRs may play a role in bridging the adaptive and innate immune responses. The TLR signalling cascade is responsible for the activation of type I interferons (IFN-α and IFN-β) in response to microbial infections 21. The first TLR to be implicated in the pathogenesis of malaria was TLR9, which signals via the type I interferon pathway 22. Taken together with our previous observations that two markers in IFNAR1 (+17470 in intron 3 and L168V in exon 4; Overall unadjusted P=0.011 and P=0.003 respectively for severe malaria) were both associated with susceptibility to P. falciparum malaria 23, we hypothesised that IFNAR1 may indeed play a role in the host response to malaria.
This study was conducted to examine the possibility that the IFNAR1 variants we previously described were in linkage disequilibrium (LD) with the causative variant(s), as the heterozygous pattern of association reported was difficult to explain in functional terms and the SNP variants themselves had no obvious functional consequences beyond a conservative amino acid change for L168V. As the region between IL10RB and IFNAR1 demonstrates a very high extent of LD we have now tested the possibility that fine-mapping using many more markers could help resolve the variant(s) involved in malaria susceptibility.
Results
A total of 45 polymorphic markers, all of them single nucleotide polymorphisms (SNPs), were genotyped and analysed across the region containing the IL10RB and IFNAR1 genes (Table 1 and Figure 1). Twenty-seven of them were novel polymorphisms identified via direct sequencing of the entire IFNAR1 gene as well as the 10 Kbp region immediately upstream of the transcription start of IFNAR1. One of the 45 polymorphisms is a non-synonymous change encoding the substitution of phenylalanine for serine at codon 8 in IFNAR2 24. Within the Gambian malaria case-control study group there was no statistically significant difference between controls and mild malaria patients when genotype and allele frequencies were compared. All control genotype frequencies were in Hardy-Weinberg Equilibrium (HWE), except for that of IL10RB +1797 (P=0.008). Additional checking with 48 randomly selected individuals revealed absolute concordance, thus minimising the chances that this could be due to genotyping error.
Table 1.
All markers genotyped in the Chr21 cytokine receptor II cluster
| Gene | Location | SNP name | dbSNP reference† |
Genotyping method |
Minor allele frequency |
Controls versus Severe Malaria cases | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Initial screen | Full screen | |||||||||
|
|
||||||||||
| Overall χ2 |
P | Overall χ2 |
P | Pcorr | ||||||
|
|
||||||||||
| IFNAR2 | exon1 | F8S | rs4986956 | Sequenom | 0.083 | 0.274 | 0.601 | |||
| IL10RB | intron2 | 224170(a-c) | rs962861 | Sequenom | 0.26 | 1.587 | 0.452 | |||
| IL10RB | intron2 | 224455(c-t) | rs2849997 | Sequenom | 0.26 | 1.04 | 0.595 | |||
| IL10RB | intron3 | 224770(g-a) | rs2285020 | Sequenom | 0.26 | 1.59 | 0.452 | |||
| IL10RB | intron3 | 225588(c-t) | rs2510317 | Sequenom | 0.26 | 0.910 | 0.635 | |||
| IL10RB | intron4 | 230146(g-a) | rs2247177 | Sequenom | 0.36 | 7.62 | 0.022 | 5.23 | 0.073 | |
| IL10RB | intron5 | 233477(g-t) | rs2850004 | Sequenom | 0.25 | 1.17 | 0.556 | |||
| IL10RB | intron6 | 242023(a-g) | rs1467845 | Sequenom | 0.20 | 0.70 | 0.706 | |||
| IL10RB | 3′UTR | +1165 | rs3171425 | Sequenom | 0.34 | 3.86 | 0.145 ¶ | 12.01 | 0.002 | 0.001 |
| IL10RB | 3′UTR | +1797 | rs1058867 | Sequenom | 0.33 | 3.55 | 0.170 ¶ | 10.18 | 0.006 | 0.010 |
| intergenic | 256042(a-c) | Sequenced | Sequenom | 0.26 | 0.72 | 0.699 | ||||
| intergenic | 260181(c-t) | Sequenced | RFLP (BglII) | 0.37 | 2.87 | 0.238 | ||||
| intergenic | 263001(a-t) | Sequenced | Sequenom | 0.24 | 2.21 | 0.332 | ||||
| IFNAR1 | 5′UTR | 264190(g-a) | Sequenced | Sequenom | 0.072 | 1.06‡ | 0.58 | |||
| IFNAR1 | 5′UTR | 264561(c-a) | Sequenced | Sequenom | 0.032 | 0.09 | 0.765 | |||
| IFNAR1 | 5′UTR | 264900(a-g) | Sequenced | Sequenom | 0.035 | 0.49 | 0.483 | |||
| IFNAR1 | 5′UTR | 267677(c-a) | Sequenced | Sequenom | 0.42 | 0.81 | 0.668 | |||
| IFNAR1 | 5′UTR | 267717(g-t) | Sequenced | Sequenom | 0.43 | 9.42 | 0.009 | 9.95 | 0.007 | 0.007 |
| IFNAR1 | 5′UTR | 268710(g-a) | Sequenced | RFLP (TaqI) | 0.34 | 4.73 | 0.091 | 6.64 | 0.036 | 0.037 |
| IFNAR1 | 5′UTR | 272354c-g | Sequenced§ | RFLP (BslI) | 0.35 | 5.75 | 0.056 | 12.39 | 0.002 | 0.007 |
| IFNAR1 | intron1 | 273628(a-g) | rs2856968 | Sequenom | 0.31 | 5.97 | 0.050 | 8.53 | 0.010 | 0.1 |
| IFNAR1 | intron1 | 273806(a-g) | rs2856969 | Sequenom | 0.33 | 6.05 | 0.049 | 5.22 | 0.073 | |
| IFNAR1 | intron1 | 274454(g-a) | rs2211687 | Sequenom | 0.24 | 0.90 | 0.639 | |||
| IFNAR1 | intron1 | 274571(a-g) | rs2226299 | Sequenom | 0.041 | 1.06‡ | 0.75 | |||
| IFNAR1 | intron1 | 275556(a-g) | rs1041429 | Sequenom | 0.32 | 0.60 | 0.741 | |||
| IFNAR1 | intron1 | 275987(c-t) | Sequenced | RFLP (BbvI) | 0.082 | 0.26‡ | 1.0 | |||
| IFNAR1 | intron1 | 276152(g-a) | Sequenced | Sequenom | 0.20 | 0.13 | 0.935 | |||
| IFNAR1 | intron1 | 276163(g-a) | Sequenced | RFLP (BtgI) | 0.30 | 1.32 | 0.516 | |||
| IFNAR1 | intron1 | 277223(t-c) | Sequenced | Sequenom | 0.069 | 0.63‡ | 0.75 | |||
| IFNAR1 | intron1 | 279350(t-c) | Sequenced | Sequenom | 0.24 | 0.38 | 0.827 | |||
| IFNAR1 | intron1 | 279749(t-c) | Sequenced | Sequenom | 0.31 | 0.95 | 0.621 | |||
| IFNAR1 | intron1 | 279923(c-t) | Sequenced | Sequenom | 0.36 | 4.25 | 0.119 ¶ | 11.65 | 0.003 | 0.004 |
| IFNAR1 | intron1 | 282792(g-a) | Sequenced | Sequenom | 0.094 | 1.54 | 0.215 | |||
| IFNAR1 | intron1 | 283455(t-g) | rs2243592 | Sequenom | 0.32 | 3.44 | 0.179 | |||
| IFNAR1 | intron2 | 283708(a-g) | rs2243594 | Sequenom | 0.35 | 9.28 | 0.0096 | 12.06 | 0.002 | 0.0003 |
| IFNAR1 | intron2 | 288443c-g | Sequenced | RFLP (BfaI) | 0.22 | 1.22 | 0.544 | |||
| IFNAR1 | exon3 | D71D | rs9981753 | RFLP (MfeI) | 0.14 | 4.34 | 0.114 ¶ | 5.79 | 0.055 | |
| IFNAR1 | intron5 | 293058(t-c) | Sequenced | RFLP (Fnu4HI) |
0.043 | 1.00‡ | 0.92 | |||
| IFNAR1 | exon8 | Thr359Met | Sequenced | RFLP (AleI) | 0.27 | 0.11 | 0.946 | |||
| IFNAR1 | intron9 | 301038(c-t) | Sequenced | Sequenom | 0.10 | 1.42‡ | 0.49 | |||
| IFNAR1 | intron10 | 302978(g-a) | Sequenced | RFLP (BsrBI) | 0.31 | 2.06 | 0.358 | |||
| IFNAR1 | 3′UTR | 304881(g-a) | Sequenced | Sequenom | 0.11 | 0.25‡ | 1.0 | |||
| intergenic | 305571(c-t) | Sequenced | RFLP (XmaI) | 0.24 | 3.09 | 0.214 | ||||
| intergenic | 309290(a-g) | Sequenced | Sequenom | 0.37 | 0.22 | 0.897 | ||||
| intergenic | 314882(g-c) | Sequenced | RFLP (BseRI) | 0.24 | 1.448 | 0.485 | ||||
All P values were derived from overall genotypic comparisons between the controls and severe malaria cases. P <0.1 are italicized; P<0.05 are in bold. Pcorr represent P values after adjusting for sickle cell trait, ethnicity, age, sex, and household location.
Those markers with a P value of <0.1 on other tests as described in materials and methods.
Denotes that the Fisher’s exact test has been performed.
Markers denoted by ‘Sequenced’ were identified by direct sequencing.
IFNAR1 272354c-g is now identified as rs2843710 on dbSNP.
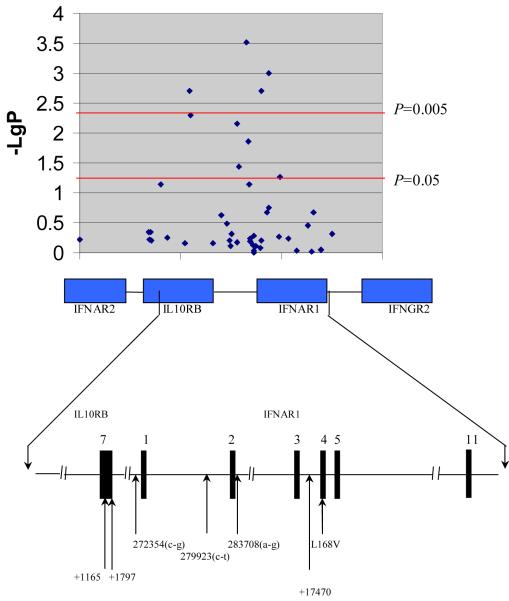
Figure 1.
High density SNP analysis showing the 45 SNPs located within the type II cytokine receptor family gene cluster on Chromosome 21 that were genotyped in the Gambian malaria study. -LgP denotes Log10P, which reflects the P values using a logarithmic scale. The bottom half of the diagram displays in greater detail the genomic organization of the region where the five markers showing the strongest evidence of association are located, together with the previously described +17470 and L168V in IFNAR1 [18].
In the initial screen, a total of 11 markers showed suggestive evidence (P<0.1) of an association with severe malaria (Table 1). Three of them (IL10RB 230146a-g, IL10RB +1165, and IL10RB +1797) were located in IL10RB, and eight of them (IFNAR1 267717g-t, IFNAR1 268710a-g, IFNAR1 272354a-g, IFNAR1 273628a-g, IFNAR1 273806a-g, IFNAR1 279923c-t, IFNAR1 283708a-g, and IFNAR1 Asp71Asp) in IFNAR1. In the full Gambian sample set, the association with markers IL10RB 230146a-g, IFNAR1 273806a-g and IFNAR1 Asp71Asp did not retain significance (P=0.073, 0.073 and 0.055 respectively). The markers for which the overall, unadjusted association remained significant in the full Gambian screening set, were further investigated by logistic regression, adjusting for the effects of possible confounders including sickle cell trait (HbAS), age (as a continuous variable), ethnicity, household location, and sex. Markers IL10RB +1165 and IL10RB +1797, as well as markers IFNAR1 267717g-t, IFNAR1 268710g-a, IFNAR1 272354c-g, IFNAR1 273628a-g, IFNAR1 279923c-t, and IFNAR1 283708a-g all remained significantly associated with severe malaria after correction (Table 1).
For markers IL10RB +1165, IL10RB +1797, IFNAR1 267717g-t, IFNAR1 272354c-g, IFNAR1 279923c-t, and IFNAR1 283708a-g, the proportion of variant genotypes (heterozygous and homozygous mutant) was found to be higher in the severe malaria cases than in the controls. Conversely, the proportion of wild-type homozygote subjects were found to be significantly under-represented in the severe malaria cases as opposed to the controls, suggesting that protection against severe malaria was conferred solely by the homozygote wild type genotype. As such, further logistic regression analyses (as previously described for sickle cell trait, age, sex, household location, and ethnic origin), using a model of dominance of the variant allele, was performed to assess the differential associations between single genotypes and severe disease phenotypes. As shown in Table 2 for the six markers mentioned above in both IL10RB and IFNAR1, the homozygous wild-type genotypes were associated with a reduced risk, and the presence of the variant alleles was associated with susceptibility to severe malaria.
Table 2.
Markers in IL10RB and IFNAR1 demonstrating evidence of association in the dominant model analysis.
| Marker | Genotype 11 versus 12 + 22§ | ||
|---|---|---|---|
| OR (95%CI) | Wald statistic | P | |
| IL10RB +1165 | 1.75 (1.25-2.50) | 10.21 | 0.001 |
| IL10RB +1797 | 1.69 (1.16-2.45) | 7.6 | 0.006 |
| IFNAR1 267717g-t | 1.39 (1.06-1.82) | 6.0 | 0.015 |
| IFNAR1 272354c-g | |||
| The Gambia | 1.5 (1.11-2.22) | 6.7 | 0.010 |
| Vietnam | 1.81 (1.17-2.82) | 7.0 | 0.005¶ |
| Kenya | 1.29 (1.00-1.66) | 4.05 | 0.022¶ |
|
| |||
| Combined populations | 1.38 (1.17-1.64) | 14.19† | 0.00017 |
|
| |||
| IFNAR1 279923c-t | 1.67 (1.18-2.38) | 8.38 | 0.004 |
| IFNAR1 283708a-g | 1.75 (1.22-2.50) | 9.39 | 0.002 |
For both Vietnamese and Kenyan replication sets, the one tailed test was used.
Mantel-Haenszel 2×2 chi-squared statistic.
11 depict the wild-type homozygote genotype, 12 depict heterozygotes, and 22 depict homozygote mutants.
N/B: The previously described IFNAR1 +17470 and L168V demonstrate P=0.018 and P=0.031 respectively in the dominant model analysis [23].
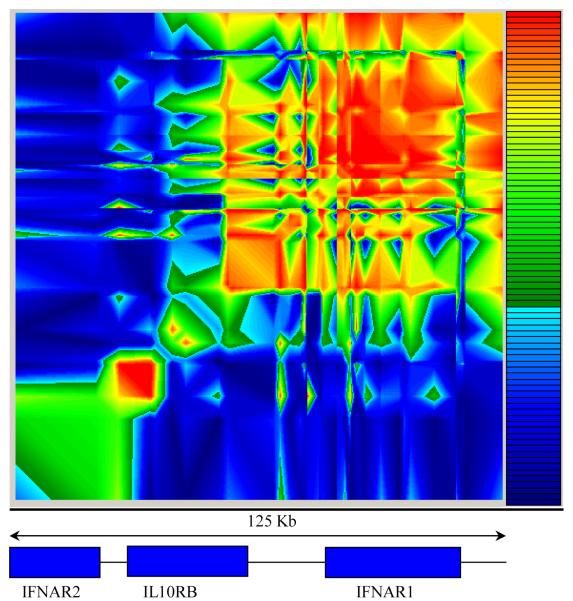
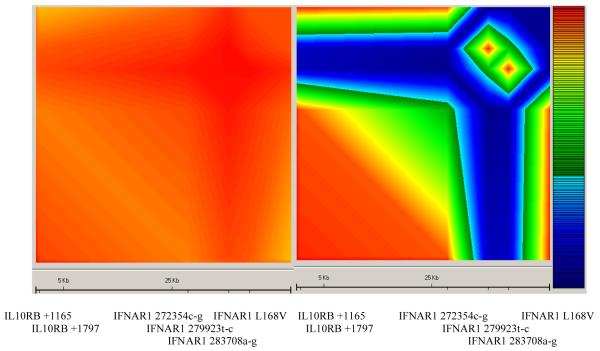
We then constructed a linkage disequilibrium (LD) map (Figure 2 and Supplementary figure A) spanning 125 kb of the Type II Cytokine Receptor Family (CRFII) gene cluster. A region of moderate LD could be distinguished, stretching approximately 50 kb from marker IL10RB +1165 to IFNAR1 L168V. The top five markers showing the strongest evidence of association with severe malaria were all confined to this region of LD. Separate LD maps were also constructed using the six most significantly associated markers in the Gambian study to compare the extent of LD between these markers in the Gambian and Vietnamese study. The comparative maps reveal that while there was very strong LD between the six markers in the Gambians, the extent of LD was much less in the Vietnamese study (Figure 3).
Figure 2.
Linkage disequilibrium (LD) results of the 45 markers in the Gambian population from GOLD (29). The key to the right shows the colour scale representing increasing LD from dark blue (D′<0.1) through to red (D′>0.9).
Figure 3.
Comparative linkage disequilibrium maps comparing the extent of LD between the Gambian (left) and Vietnamese (right) studies constructed using the six polymorphisms found to be highly significant in the Gambian malaria study. The key to the right shows the colour scale representing increasing LD from dark blue (D′<0.1) through to red (D′>0.9).
Haplotypes were reconstructed using the data from the 6 markers (5 from this study, and the previously described IFNAR1 L168V [23]) showing the strongest association with severe malaria (IL10RB +1165, IL10RB +1797, IFNAR1 272354c-g, IFNAR1 279923c-t, IFNAR1 283708a-g, as well as IFNAR1 L168V) which were shown to be in strong LD. Two main haplotypes were observed: IL10RB +1165-A IL10RB +1197-G IFNAR1 272354-C IFNAR1 279923-C IFNAR1-283798-A IFNAR1 L168V-G and IL10RB +1165-G IL10RB +1797-A IFNAR1 272354-G IFNAR1 279923-T IFNAR1 283708-G IFNAR1 L168V-C. No significant difference in haplotype frequencies was observed between the controls and severe malaria cases (PHASE2 Global test for significance P=0.66).
Two chromosome haplotype combinations (also known as diplotypes, previously described 23) were then constructed using the haplotypes obtained from PHASE2. The ‘homozygous’ AGCCAG/AGCCAG diplotype was found to be more prevalent in controls compared to the cases, and conversely, the ‘heterozygous’ AGCCAG/GAGTGC diplotype was found to be over represented in the severe malaria cases compared to the controls (3×2 χ2=12.17, P=0.002, Supplementary table B). However, this result observed with the diplotypes was no more informative than the analysis of the single markers.
We then attempted to replicate the associations of the five most significantly associated markers (IL10RB +1165, IL10RB +1797, IFNAR1 272354c-g, IFNAR1 279923c-t, IFNAR1 283708a-g, as well as IFNAR1 L168V) in an independent replication set using Vietnamese severe malaria cases and controls. Only for IFNAR1 272354c-g was a significant association detected, with the C/C homozygote wild-type genotype significantly under-represented among severe malaria cases (31.5%) (2×2 χ2 = 6.68, P=0.0097, Pone tailed=0.0049) compared to controls (44.9%). The overall 3×2 χ2 test involving all three genotypes was also significant (Supplementary table A), and was unchanged by allowance for Haemoglobin E (HbE) genotype. In an analysis of alleles, the wild-type C allele was significantly under-represented in the cases with severe malaria (55.3%) (2×2 χ2 = 5.88, P=0.015) compared to the controls (64.5%).
We then attempted to replicate the observed association with IFNAR1 272354c-g in a third population from Kilifi, Kenya. A significant difference in overall genotype frequencies was observed between the severe malaria cases and controls (3×2 χ2=6.24, P=0.044; Supplementary table A). Further analysis revealed a lower proportion of homozygous wild-type C/C genotype individuals among severe malaria cases (46.8%) compared to controls (52.1%)(2×2 χ2=4.05, P=0.044, Pone tailed=0.022).
A combined analysis involving the Gambian, Kenyan and Vietnamese studies was then performed using the Mantel-Haenszel 2×2 χ2 test stratified for the three different populations. The results demonstrate a highly significant under-representation of the C/C wild type homozygote genotypes in the severe malaria cases compared to the controls, and conversely an over-representation of the variant G allele in the severe malaria cases (N=2444, Mantel-Haenszel 2×2 χ2 = 14.19, P=1.7×10−4; OR=1.38, 95% CI= 1.17-1.64).
Discussion
In this study, we have shown a clear pattern of association, in which protection was afforded by the homozygote wild-type genotype, and susceptibility conferred by the presence of the variant allele and this was observed for five polymorphic markers across a 50 Kb region in the Gambian study. These markers, which were in strong linkage disequilibrium (Figure 3) in the Gambia, demonstrate highly significant associations with severe malaria, and this remains after correction for the potential confounding effects of sickle cell trait, ethnicity, age, household location, and sex. It is very likely that the functional variant lies within this 50Kb region because it is contained within a well-defined block of high linkage disequilibrium (Figure 2 and Supplementary figure A). The reconstruction of haplotypes and diplotypes using the data obtained from the most significantly associated markers from the Gambian study failed to yield further information in resolving the functional variant. This is probably due to the fact that all 5 markers (together with IFNAR1 L168V) were in such strong LD. No other association was detected 3′ of IFNAR1 L168V even after typing an additional eight markers downstream of it. The significant association between IFNAR1 272354c-g and protection against severe malaria was replicated in an independent Vietnamese population, and a borderline significant result observed in the study involving Kenyan children. These observations considerably lessen the likelihood that the initial observed association had occurred by chance. Apart from IFNAR1 272354c-g, the associations with the other markers were not replicated in the Vietnamese population, further increasing the likelihood that this marker, if it is not the functional polymorphism is at least in very strong linkage disequilibrium with it. We have also shown that the extent of LD between the highly significant markers in the Gambian study is very strong (Figure 3), suggesting that the other markers were found to be significantly associated because they are in strong LD with IFNAR1 272354c-g. On the other hand, the extent of pairwise LD between the above-mentioned markers was found to be less pronounced in the Vietnamese population (Figure 3), consistent with the failure of replication for the other five SNPs in the Vietnamese study. IFNAR1 272354c-g is 576 base pairs away from the transcription start site, and as promoter and binding sites for regulatory elements are often found up to 1000 base pairs upstream of the transcription start site, this polymorphism could well affect the expression levels of IFNAR1. Further functional studies on the effect of promoter variants on IFNAR1 transcript levels are therefore merited.
The positive results presented here have not been corrected for multiple-testing using the Bonferroni correction. This assumes completely random selection of markers across a random selection of genes. However, most of the markers in IL10RB and IFNAR1 have been shown to be in strong linkage disequilibrium, and hence are not independent of one another. This, together with the successful replication of the IFNAR1 272354c-g association in two further populations, reduces considerably the possibility that this was a chance finding. Even if the conservative Bonferroni correction (taking into consideration all 45 markers genotyped) were to be applied to the unadjusted overall association observed with IFNAR1 272354c-g (P=1.7×10−4), the final corrected P value remains significant at P=0.007.
Although there is no direct evidence linking type-1 interferons to malaria pathogenesis, reports have linked IFN-γ and the efficacy of clearance of liver stage P. falciparum 25, 26. There are studies 17, 18 which describe a possible signalling connection between IFN-α and IFN-γ, where IFN-α is thought to act upstream of IFN-γ. This further underpins the importance of type-I interferons in the pathogenesis of severe malaria and IFNAR1 as a functional candidate in this disease. This detailed genetic analysis suggests that further study of the role of type I interferons in malaria pathogenesis is warranted, and also underlines the value of independently recruited replication sets in helping to identify functional variants in genomic regions where the LD between markers is strong.
Patients and methods
Subjects
The details for the Gambian malaria case-control study have been described previously 27. The Vietnamese subjects were adults with severe malaria admitted to the Centre for Tropical Diseases, Ho Chi Minh City, Vietnam recruited between 1991 and 1996. Controls were ethnically matched adult non-malaria cases recruited from other departments of the same hospital. Subjects with mild malaria were not included in this study. Ethical approval was granted by the Ethical and Scientific committee of the Centre for Tropical Diseases in Ho Chi Minh City, Vietnam.
For the Kenyan study, children admitted to the high dependency unit of Kilifi District Hospital with severe malaria 28 were recruited as cases. The controls were children who had been recruited from the community surrounding the hospital for a study of bacteraemia, matched by age, sex and household location to children admitted to the hospital with bacteraemia. All samples were obtained only after informed consent and permission from a parent or guardian. Ethical approval for this study was granted by the Kenya Medical Research Institute (KEMRI) National Scientific Steering and Research Committees.
Identification of polymorphisms
SNP markers were identified via direct sequencing of 24 Gambian children (12 cases and 12 controls), as well as obtained via dbSNP database searches. Individual marker details are included in Table 1. For the sequencing exercise, a 44 Kbp genomic region (comprising the entire coding region of IFNAR1, most of the non-coding intronic sequence, a region 10Kbp upstream of the transcription start, as well as a region 4 Kbp downstream of the terminal exon) was subjected to direct DNA sequencing (primers appended in Supplementary table C). The genomic regions of interest were first amplified by PCR, followed by direct sequencing using BigDye v3.1 (Applied Biosystems). The products were analysed using an ABI 3700 capillary sequencer and the trace files computed via poly PhredPhrap 29, 30 and visualised using Consed 31.
Screening strategy
A detailed description of the screening strategy can be found in supplementary information.
Genotyping methods
Most of the markers were genotyped using the Sequenom MassArray primer extension method 32. Other details on genotyping (including the primers used) are presented in Supplementary Information and Supplementary table D.
Statistical analysis
Initially, the markers were analyzed individually. The Pearson’s χ2 test was used to compare the differences in allele frequencies and the relevant overall χ2 test (2×2 or 3×2) for differences in genotype distribution between the cases and controls using SPSS version 12.0 (SPSS Inc.). We then used forward stepwise logistic regression, including variables that were significantly associated with malaria on univariate analysis (P<0.05) to assess the overall association between genetic markers adjusted for the potential confounding effects of the variables sickle cell trait status, age, sex, ethnicity, and household location.
As both the Vietnamese and Kenyan studies are replication sets, one-tailed tests of significance were used for 2×2 comparisons. This is justified because there is an a priori knowledge of the direction of the association. When an association was successfully replicated in two or more populations, the most consistent model (dominance, recessive, or allelic) was adopted, and the data from the relevant populations was then combined and analyzed as a whole, stratifying for the different study populations using the Mantel-Haenszel 2×2 χ2 test (SPSS version 12.0). In the Gambian malaria study, there were no statistically significant differences between the mild, non-malaria controls and the severe, non-malaria controls for any genotype or allele frequency, so these control subjects were pooled together as a single control group for subsequent analysis.
Haplotype analysis
When associations were observed between one or more genetic markers and severe malaria, the marker data were used for haplotype reconstruction. The haplotypes were reconstructed from genotype data using the PHASE2 package 33, which is designed for haplotype estimation from case control studies. PHASE2 conducts a global test of significance between cases and controls, while accounting for haplotype uncertainty. The combination of haplotypes for each individual (e.g. the diplotype) was analysed using Pearson’s χ2 test.
Linkage disequilibrium analysis
Analysis of pairwise linkage disequilibrium between markers was conducted using the Graphical Overview of Linkage Disequilibrium (GOLD) package 34.
Acknowledgements
The authors would like to thank the patients from the Gambian, Kenyan, and Vietnamese malaria study populations, as well as the many investigators involved in the original case-control studies in these populations for their contributions. The authors gratefully acknowledge Angela Frodsham and Branwen Hennig for information on IFNAR2 F8S and IL10RB markers +1165 and +1797.
Funding This work was funded by the Wellcome Trust, UK and the Agency for Science, Technology and Research (A-STAR), Singapore. CCK is a scholar of A-STAR and is a member of the MBBS-PhD programme, Faculty of Medicine, National University of Singapore. AS, JAB, KM, NP, KM, and TNW are supported by the Wellcome Trust and the Kenya Medical Research Institute. AVSH is a Wellcome Trust Principal Research Fellow. This study was published with permission from the Director of KEMRI.
Footnotes
Competing interests The authors declare no competing financial interests.
Supplementary information is available at the Genes and Immunity website’
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434(7030):214–7. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison AC. Polymorphism and natural selection in human populations. Cold Spring Harb Symp Quant Biol. 1964;29:137–49. doi: 10.1101/sqb.1964.029.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, Gupta S, et al. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 1995;376(6537):246–9. doi: 10.1038/376246a0. [DOI] [PubMed] [Google Scholar]
- 4.Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321(6072):744–50. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 5.Hutagalung R, Wilairatana P, Looareesuwan S, Brittenham GM, Aikawa M, Gordeuk VR. Influence of hemoglobin E trait on the severity of Falciparum malaria. J Infect Dis. 1999;179:283–286. doi: 10.1086/314561. [DOI] [PubMed] [Google Scholar]
- 6.Hutagalung R, Wilairatana P, Looareesuwan S, Brittenham GM, Gordeuk VR. Influence of hemoglobin E trait on the antimalarial effect of artemisinin derivatives. J Infect Dis. 2000;181:1513–1516. doi: 10.1086/315373. [DOI] [PubMed] [Google Scholar]
- 7.Frodsham AJ, Hill AV. Genetics of infectious diseases. Hum Mol Genet. 2004;13 Spec No 2:R187–94. doi: 10.1093/hmg/ddh225. [DOI] [PubMed] [Google Scholar]
- 8.Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, et al. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet. 1999;22(2):145–50. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 9.McGuire W, Knight JC, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Severe malarial anemia and cerebral malaria are associated with different tumor necrosis factor promoter alleles. J Infect Dis. 1999;179(1):287–90. doi: 10.1086/314533. [DOI] [PubMed] [Google Scholar]
- 10.Koch O, Awomoyi A, Usen S, Jallow M, Richardson A, Hull J, et al. IFNGR1 gene promoter polymorphisms and susceptibility to cerebral malaria. J Infect Dis. 2002;185(11):1684–7. doi: 10.1086/340516. [DOI] [PubMed] [Google Scholar]
- 11.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39(1):113–9. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 12.Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39(1):120–5. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39(1):126–30. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 14.Hugosson E, Montgomery SM, Premhi Z, Troye-Blomberg M, Bjorkman A. Higher IL-10 levels are associated with less effective clearance of Plasmodium falciparum parasites. Parasite Immunol. 2004;26(3):111–7. doi: 10.1111/j.0141-9838.2004.00678.x. [DOI] [PubMed] [Google Scholar]
- 15.Lyke KE, Burges R, Cissoko Y, Sangare L, Dao M, Diarra I, et al. Serum levels of the proinflammatory cytokines interleukin-1 beta (IL-1beta), IL-6, IL-8, IL-10, tumor necrosis factor alpha, and IL-12(p70) in Malian children with severe Plasmodium falciparum malaria and matched uncomplicated malaria or healthy controls. Infect Immun. 2004;72(10):5630–7. doi: 10.1128/IAI.72.10.5630-5637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vigario AM, Belnoue E, Cumano A, Marussig M, Miltgen F, Landau I, et al. Inhibition of Plasmodium yoelii blood-stage malaria by interferon alpha through the inhibition of the production of its target cell, the reticulocyte. Blood. 2001;97(12):3966–71. doi: 10.1182/blood.v97.12.3966. [DOI] [PubMed] [Google Scholar]
- 17.Cousens LP, Peterson R, Hsu S, Dorner A, Altman JD, Ahmed R, et al. Two roads diverged: interferon alpha/beta- and interleukin 12-mediated pathways in promoting T cell interferon gamma responses during viral infection. J Exp Med. 1999;189(8):1315–28. doi: 10.1084/jem.189.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000;288(5475):2357–60. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 19.Hunt NH, Grau GE. Cytokines: accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 2003;24(9):491–9. doi: 10.1016/s1471-4906(03)00229-1. [DOI] [PubMed] [Google Scholar]
- 20.John CC, Moormann AM, Sumba PO, Ofulla AV, Pregibon DC, Kazura JW. Gamma interferon responses to Plasmodium falciparum liver-stage antigen 1 and thrombospondin-related adhesive protein and their relationship to age, transmission intensity, and protection against malaria. Infect Immun. 2004;72(9):5135–42. doi: 10.1128/IAI.72.9.5135-5142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–7. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 22.Pichyangkul S, Yongvanitchit K, Kum-arb U, Hemmi H, Akira S, Krieg AM, et al. Malaria blood stage parasites activate human plasmacytoid dendritic cells and murine dendritic cells through a Toll-like receptor 9-dependent pathway. J. Immunol. 2004;172(8):4926–33. doi: 10.4049/jimmunol.172.8.4926. [DOI] [PubMed] [Google Scholar]
- 23.Aucan C, Walley AJ, Hennig BJ, Fitness J, Frodsham A, Zhang L, et al. Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun. 2003;4(4):275–82. doi: 10.1038/sj.gene.6363962. [DOI] [PubMed] [Google Scholar]
- 24.Frodsham AJ, Zhang L, Dumpis U, Azizah N, Best S, Durham A, et al. A Class II cytokine Receptor Gene Cluster is a major locus for Hepatitis B persistence. Proc Natl Acad Sci U S A. 2006;103(24):9148–53. doi: 10.1073/pnas.0602800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolan DL, Sedegah M, Hedstrom RC, Hobart P, Charoenvit Y, Hoffman SL. Circumventing genetic restriction of protection against malaria with multigene DNA immunization: CD8+ cell-, interferon gamma-, and nitric oxide-dependent immunity. J Exp Med. 1996;183(4):1739–46. doi: 10.1084/jem.183.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luty AJ, Lell B, Schmidt-Ott R, Lehman LG, Luckner D, Greve B, et al. Interferon-gamma responses are associated with resistance to reinfection with Plasmodium falciparum in young African children. J Infect Dis. 1999;179(4):980–8. doi: 10.1086/314689. [DOI] [PubMed] [Google Scholar]
- 27.Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352(6336):595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 28.Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332(21):1399–404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 29.Ewing Brent, Hillier LaDeana, Wendl Michael C., Green Phil. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 30.Ewing Brent, Green Phil. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 31.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 32.Jurinke C, van den Boom D, Cantor CR, Koster H. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 33.Stephens M, Smith NJ, Donnelly P. new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis GR, Cookson WO. GOLD- Graphical Overview of Linkage Disequilibrium. Bioinformatics. 2000;16(2):182–3. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]