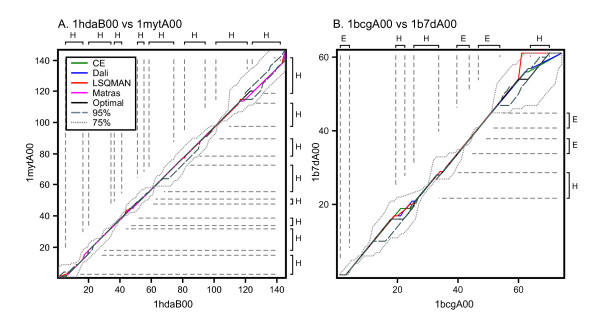
Figure 1.
Alignment paths of structure-based, and optimal and suboptimal sequence alignments. Two pairs of aligned proteins are shown: (A) 1 hdaB00 vs 1 mytA00, pair 25 in Figure 4, 26.8% identity, E() 2.1 × 10-10; and (B) 1bcgA00 vs. 1b7dA00, pair 42 in Figure 4, 33.3% identity, E() = 9.6 × 10-7. Four structural alignments are shown: CE (green), DALI (blue), LSQMAN (red), and Matras (magenta). Also shown is the best global sequence alignment (black, scoring matrix BLOSUM50, gap penalties -10/-2), the envelope of alignments within 95% of optimal (grey, dashes), and the envelope of alignments within 75% of optimal (light grey, dots). The Zuker suboptimal alignment algorithm produced 2 optimal alignments for 1 hdaB00:1 mytA00, 45 alignments at 95% optimal, and 1,170 at 75% optimal. For 1bcgA00:1b7dA00 there were 8 optimal, 34 95% optimal, and 349 75% optimal alignments produced. Both protein pairs are from the medium sequence similarity group. The axes and vertical/horizontal dashed gray lines highlight the secondary structure elements as defined by the PDB file.