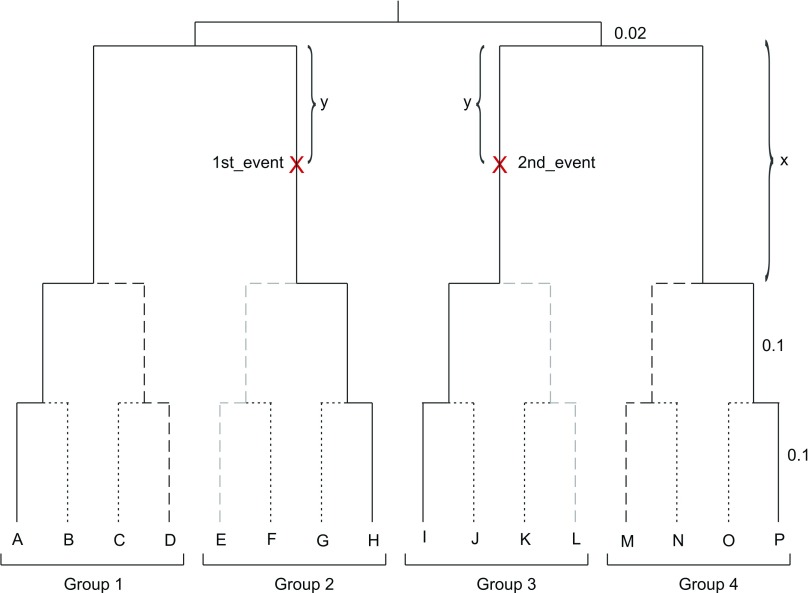
FIGURE 1.
Simulations were done on a 4-taxon tree: T4=((A,H),(I,P)) (solid lines), two 6-taxon trees: (A,(E,H)),((I,L),P)) (solid and light dashed lines) and ((A,D),H),(I,(M,P))) (solid and dark dashed lines), an 8-taxon tree: (A,D),(E,H)),((I,L),(M,P))) solid and both light and dark dashed lines, and a 16-taxon tree: T16=((((A,B),(C,D)),((E,F),(G,H))),(((I,J),(K,L)),((M,N),(O,P)))) (all lines). At the root, 80% of the sites were set as invariable. Pvar+=(0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50) percent of the invariable sites were reset to be variable in 2 events marked as “1st_event” and “2nd_event.”