Figure-3. Nuclear and cytoplasmic sense–antisense RNA pairing.
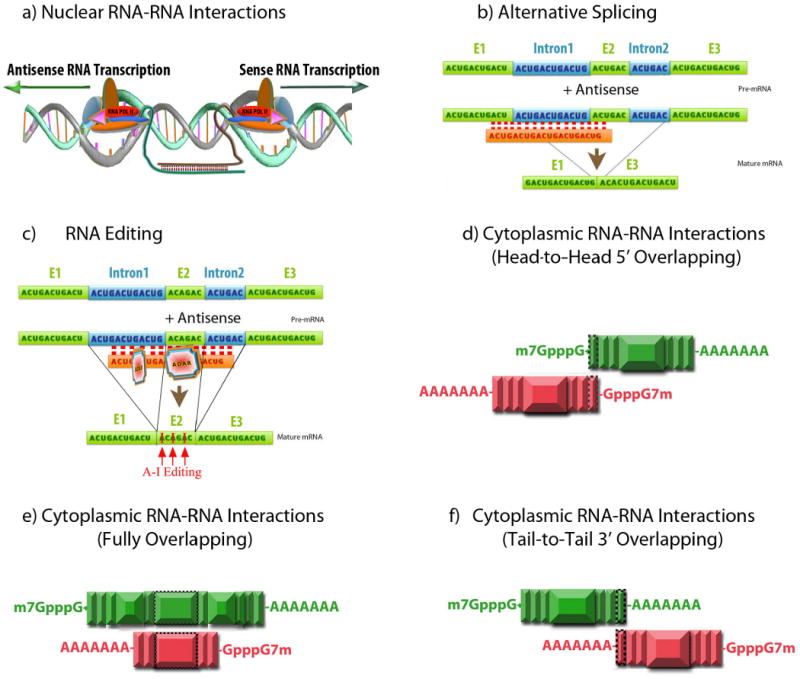
(Aa) Nuclear RNA duplex formation might occur locally after transcription, thereby inhibiting sense RNA processing. The RNA processing includes capping, polyadenylation, nuclear localization and transport, which all might be affected by nuclear sense-antisense RNA duplex formation. (Ab) Natural antisense transcripts can cover donor and acceptor splice sites in the sense pre-mRNA transcript to change alternative splicing patterns. (Ac) RNA editing is another possible consequence of nuclear RNA duplex formation, which causes A-to-I editing by the ADAR (adenosine deaminases that act on RNA) enzyme. RNA editing can induce nuclear retention and degradation of hyper-edited transcript by inosine-specific nucleases or alternatively, editing can result in alteration of the amino acid sequence. (B) Cytoplasmic sense–antisense duplex formation can occur in three different formats; (Ba) head-to-head 5′ overlapping, (Bb) fully overlapping in which antisense is embedded in the sense transcript or (Bc) tail-to-tail 3′ overlapping configuration. Exons are depicted as vertical boxes and the overlapping region is shown with a border. Cytoplasmic sense–antisense RNA duplex formation can possibly exert its own effects on sense mRNA. Hiding or exposing AU-rich elements in sense transcript can affect RNA stability. Changes in the RNA secondary structure, after binding to antisense RNA, can alter translation, subcellular localization and accessibility of the RNA degradation machinery. According to this model, natural antisense transcripts can potentially ‘mask’ miRNA-binding sites and release the miRNA-induced block of translation. Finally, Dicer can also process sense–antisense RNA duplexes to endogenous siRNAs.
