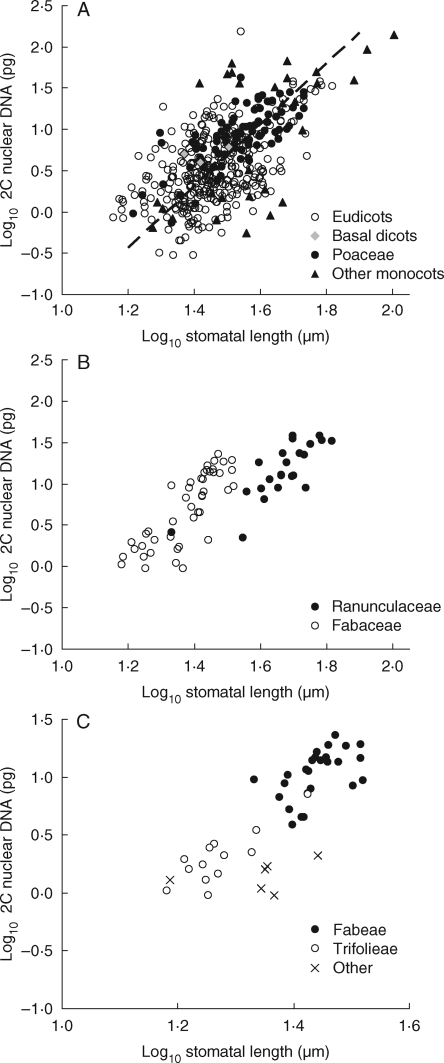
Fig. 5.
Examples of the relationship between stomatal and genome size. (A) All species: r2 = 0·36, P < 0·001, n = 446. Eudicots (r2 = 0·26, P < 0·001, n = 326); basal dicots (n = 3); monocots, excluding Poaceae (r2 = 0·39, P < 0·001, n = 30); and Poaceae (r2 = 0·53, P < 0·001, n = 87), as indicated. (B) Contrasted families: Ranunculaceae (r2 = 0·64, P < 0·001, n = 23), and Fabaceae (r2 = 0·64, P < 0·001, n = 46), as indicated. Other families: Asteraceae (r2 = 0·08, P < 0·05, n = 51; minus Chrysanthemum segetum, r2 = 0·08); Caryophyllaceae (r2 = 0·23, P < 0·1, n = 15); Polygonaceae (r2 = 0·32, P < 0·05, n = 13). (C) Contrasted tribes (Fabaceae): Fabeae (r2 = 0·29, P < 0·01, n = 26); Trifolieae (r2 = 0·67, P < 0·001, n = 13); and other (n = 7), as indicated. Tribes in other families: Asteraceae, Anthemideae (r2 = 0·13, n.s., n = 17; minus Chrysanthemum segetum, r2 = 0·30, P < 0·05, n = 16); Lactuceae (r2 = 0·50, P < 0·001, n = 18); Poaceae, Agrostideae (r2 = 0·49, P < 0·01, n = 13); Aveneae (r2 = 0·72, P < 0·001, n = 11); Poeae (r2 = 0·05, n.s., n = 19).