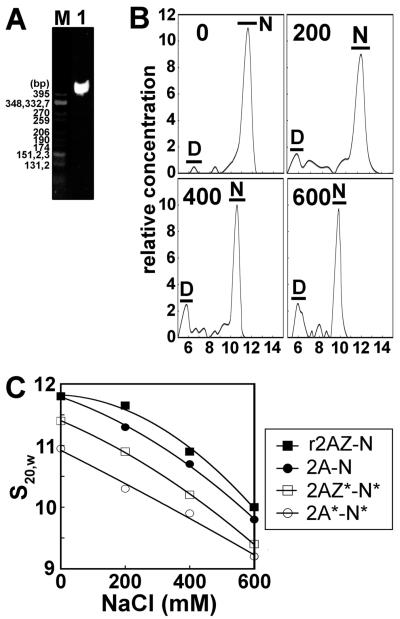
Figure 3.
Sedimentation velocity analysis of acetylated nucleosomes. (A) Native PAGE of nucleosome core particles reconstituted with 155 bp random sequence chicken erythrocyte DNA and histone octamers consisting of native purified H2A and native non-acetylated histone complement. The marker is CfoI-digested pBR322 plasmid DNA. (B) Profile of analytical ultracentrifuge analysis of nucleosomes containing H2A and a native non-acetylated histone complement. The graphs show plots of the relative sample concentration vs the sedimentation coefficient at four different NaCl concentrations (0, 200, 400, and 600 mM). Data were obtained using the histogram envelope analysis from UltraScan (see Materials and Methods). D: free 155 bp DNA. N: nucleosome core particle. (C) Ionic strength dependence of the sedimentation coefficient (s20,W) of reconstituted NCPs that were dialyzed against 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA buffer containing different NaCl concentrations and analyzed by sedimentation velocity at 44000 rpm and 20 °C.