Figure 2. CXCR4 transcription was analyzed in colorectal tumor cell lines and primary tissue by Reverse Transcription Polymerase Chain Reaction (PCR).
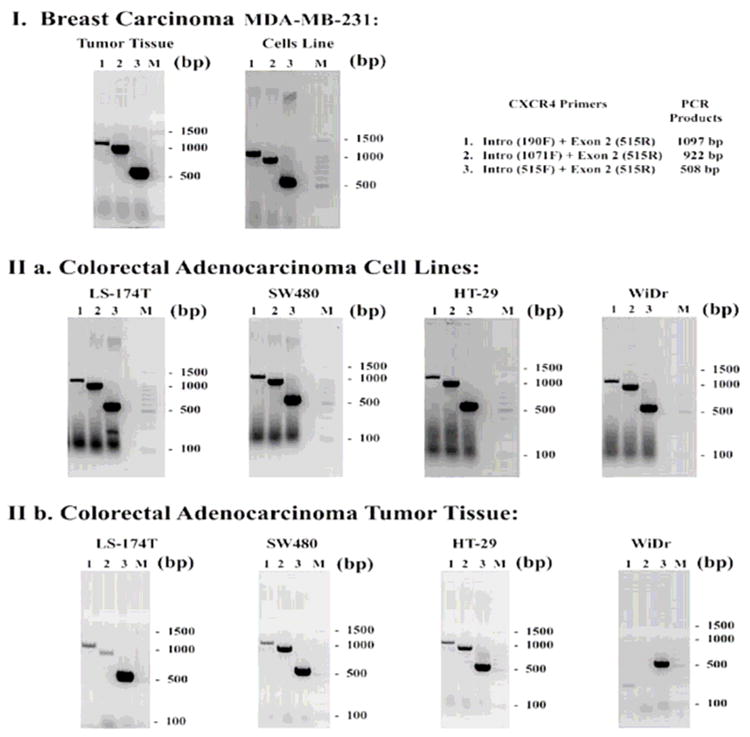
RNA was isolated from cells or from tumor tissue and reverse transcribed. The resultant PCR products were fractionated on a 1.5% agarose gel followed by ethidium bromide staining, and then analyzed by ABI Prism 3130 xI Sequence Detection System and 3130 xI Genetic Analyzer data collection software v 3.0. Lane M: 100 bp DNA ladder; lane 1: 1097 bp CXCR4 band; lane 2: 922 bp CXCR4 band; lane 3: 508 bp CXCR4 band. (I) As a positive control for RNA expression of CXCR4, a breast cancer cell line, MDA-MB-231, and in breast tumor tissue were analyzed. (II) Four colorectal adenocarcinoma cell lines (LS-174T, SW480, HT-29, WiDr) were analyzed for CXCR4 transcription. (III) Tumor xenografts grown subcutaneously from injected cells were also analyzed for CXCR4 transcription. The PCR products were designed to determine whether the CXCR4 transcript has deletions in it (e.g., IIb, WiDr appears to be a truncated transcript).
