FIGURE 3.
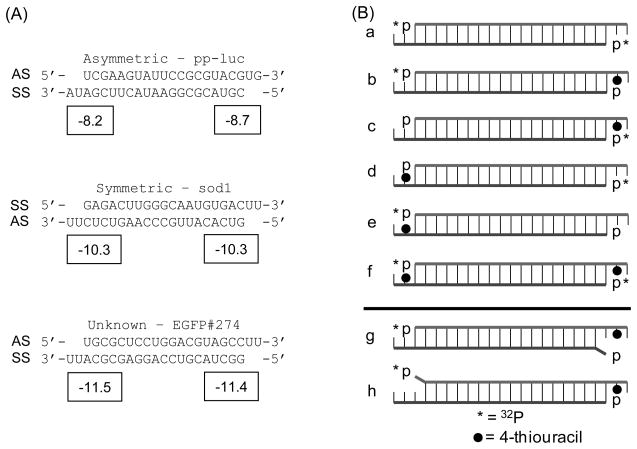
Nucleic acids used. (A) Three siRNA sequences were used in this study. Two have been documented previously for their asymmetric (pp-luc; referred to in the text as sequence ‘A’) or symmetric (sod1; sequence ‘S’) silencing activities and recognition by Dicer-2/R2D2 in Drosophila (7, 34). The third was previously used for EGFP silencing (EGFP; sequence ‘G’) and was expected to be slightly asymmetric based on silencing activity (40) and ΔΔG calculation with mfold (57). AS denotes the antisense strand and SS denotes the sense strand. Boxed numbers indicate ΔG values (in kcal/mol) calculated from the four terminal nearest-neighbor base-pairs, including the A:U penalty and 3′-overhang contribution (single base stacking energy). (B) A series of 8 different siRNAs were created for each sequence as described in the text. The strand location in (A) matches the strand location in (B). * denotes strands 5′-labeled with 32P-ATP; ● denotes 4-thiouracil modification at position 20; siRNAs ‘g’ and ‘h’ contain a single 5′-terminal mismatch as indicated by the protruding base.