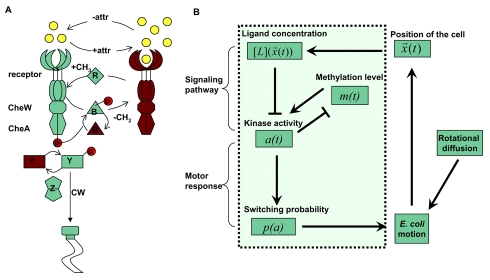
Figure 1. Illustrations of the E. coli chemotaxis pathway and the SPECS (Signaling Pathway-based E. coli Chemotaxis Simulator) model.
(A) The E. coli chemotaxis signal transduction pathway. The MCP complex receptor-CheW-CheA is the sensor and can be active (green) or inactive (dark brown). Binding of attractant molecules (yellow) decreases the probability of the receptor to be active. Once activated, the histidine kinase CheA quickly autophosphorylates and then transfers the phosphate group to CheY. CheY-p, the response regulator, diffuses to the flagellar motor and modulates its switching probability between CW and CCW rotations. CheZ, the CheY-p phosphatase, serves as the sink of the signal. Adaptation of the system is carried out by methylation and demethylation of the receptor, which are facilitated by the enzymes, CheR and CheB-p, respectively. (B) Flow chart of the SPECS model (reproduced from Figure 3 in [21]). The input for the signaling pathway (inside the box) is the instantaneous ligand concentration 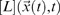 . The internal signaling pathway dynamics is described at the coarse-grained level by the interactions between the average receptor methylation level
. The internal signaling pathway dynamics is described at the coarse-grained level by the interactions between the average receptor methylation level 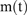 and the kinase activity
and the kinase activity 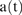 , which eventually determines the switching probability of the flagellar motor
, which eventually determines the switching probability of the flagellar motor 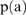 . The switching probability is then used to determine the cell motion (run or tumble), and direction of motion during run fluctuates due to rotational diffusion. The motion of the cell leads it to a new location with a new ligand concentration for the cell and the whole simulation process continues.
. The switching probability is then used to determine the cell motion (run or tumble), and direction of motion during run fluctuates due to rotational diffusion. The motion of the cell leads it to a new location with a new ligand concentration for the cell and the whole simulation process continues.