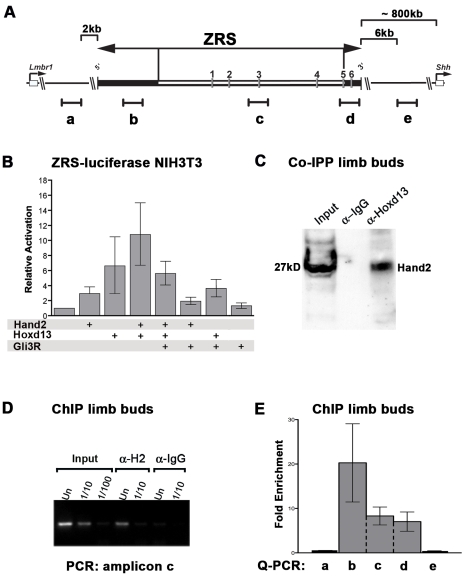
Figure 4. Hand2 interacts with Hoxd13 and is part of the chromatin complexes bound to the ZRS in limb buds.
(A) Scheme of the ∼1.7 kb mouse ZRS cis-regulatory and the flanking genomic regions. The ZRS is located within an intron of the mouse Lmbr1 gene (indicated on the left) and located ∼800 kb upstream of the Shh proximal promoter and coding exons (indicated on the right, see also Figure S5). The evolutionary conserved ZRS region drives expression of a LacZ reporter gene in Shh-like pattern in mouse limb buds [28], while deletion of the MFCS1 core region (indicated in white) disrupts Shh activation in limb buds [29]. Six Ebox sequences in the ZRS, which could potentially interact with Hand2 proteins are numbered “1” to “6”. Black lines indicate the approximate positions and sizes of the PCR amplicons for ChIP analysis. Note that amplicons “b” to “d” reside within the mouse ZRS, while amplicons “a” and “e” are located ∼2 kb upstream and ∼6 kb downstream of the ZRS and serve as non–ZRS controls. (B) Luciferase transactivation assay in NIH3T3 fibroblasts. Cells were co-transfected with ZRS-Luc and the expression plasmids indicated. Bars represent standard deviations. P<0.0001 for all samples except Gli3R alone: P = 0,0519. (C) Co-immunoprecipitation of Hand2 and Hoxd13 from wild-type limb buds (E10.5) using anti-Hoxd13 antibodies (α-Hoxd13) or IgGs (control). Hand2 proteins associated to Hoxd13 protein complexes were detected by Western blotting. (D,E) ChIP from wild-type limb buds (E11.0) to detect Hand2-containing chromatin complexes bound to the ZRS. (D) Analysis of amplicon “c” by conventional PCR (186 bp). Input: DNA isolated from cross-linked chromatin of E11.0 limb buds prior to ChIP was used as a positive control for PCR amplification. α-H2: ChIP using Hand2 antibodies. α-IgG: ChIP using non-specific goat IgGs as a control. Un: undiluted sample; dilutions as indicated. (E) Q–PCR analysis of three completely independent ChIP experiments using freshly cross-linked chromatin and α-Hand2 antibodies. The average values ± standard error are shown. Values obtained by amplifying a particular region from ChIP experiments using non-specific goat IgGs were arbitrarily set at 1 and used to calculate the values for the α-Hand2 ChIP experiments. Statistical evaluation by the Mann-Whitney test shows that the amplicons within the ZRS (“b” to “d”) are enriched in a statistically highly significant manner in comparison to the adjacent non-ZRS amplicons (“a” and “e”; p = 0.0018).