Fig. 1.
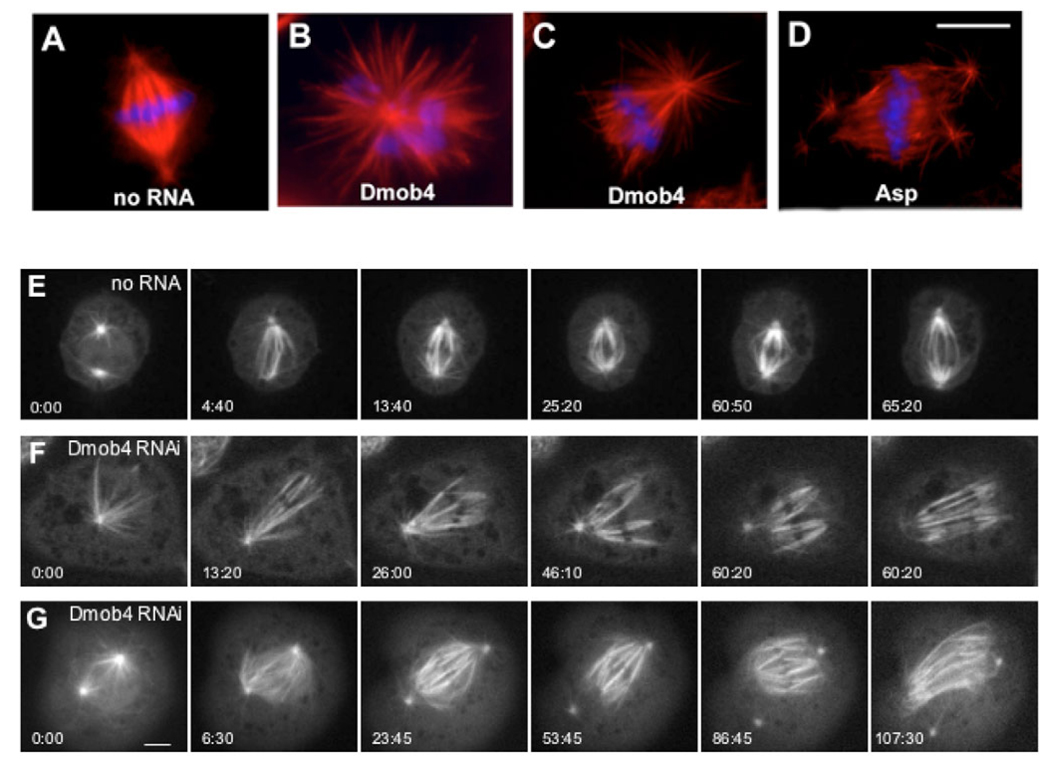
Mitotic phenotypes observed after RNAi knockdown of Drosophila Mob4 (Dmob4) in S2 cells. Cells were treated with dsRNA and were fixed and stained with antibodies against tubulin (red) and with DAPI (blue) at day 4 (see Materials and Methods). (A) Untreated cells display bipolar spindles with well-defined centrosomes. (B) Mob4 RNAi cells show abnormally high numbers of monopolar spindles (32%, data not shown) as well as monastral bipolar spindles with splayed kinetochore minus ends (41%, data not shown). This latter defect resembled the frayed spindles that formed after Asp RNAi (D). Bar, 10 µm (A–D). Live imaging of GFP-tubulin revealed further details of the defects in spindle formation arising from Mob4 depletion (E–G). (E) Control cell (no RNA) with two centrosomes forming a bipolar spindle with spindle fibers organized normally at the poles. Time (in minutes and seconds) from the first frame is indicated in the lower left corner of each panel. The rightmost panel shows a cell in early anaphase (same in F and G), indicating the time of anaphase onset. (Full time-lapse movie available in supplementary material Movie 1.) (F) This Mob4-RNAi-treated cell initially formed a monopolar spindle that then converted to a monastral bipolar spindle. The acentrosomal pole on the right never becomes organized, and the left pole loses focus when the centrosome and K fibers begin to dissociate in prometaphase (4th panel). (Full time-lapse movie available in supplementary material Movie 2.) (G) Mob4-RNAi-treated cell with two centrosomes forming a bipolar spindle that becomes unorganized at both poles owing to detachment of K fibers from the centrosome and splaying during prometaphase/metaphase. (Full time-lapse movie available in supplementary material Movie 3.) Anaphase onset in Mob4-depleted cells occurs within the same time range observed in control cells. The K fiber splaying observed after Mob4 depletion resembles that described in live-cell imaging of cells depleted of the Asp protein (data not shown). Bar, 5 µm.