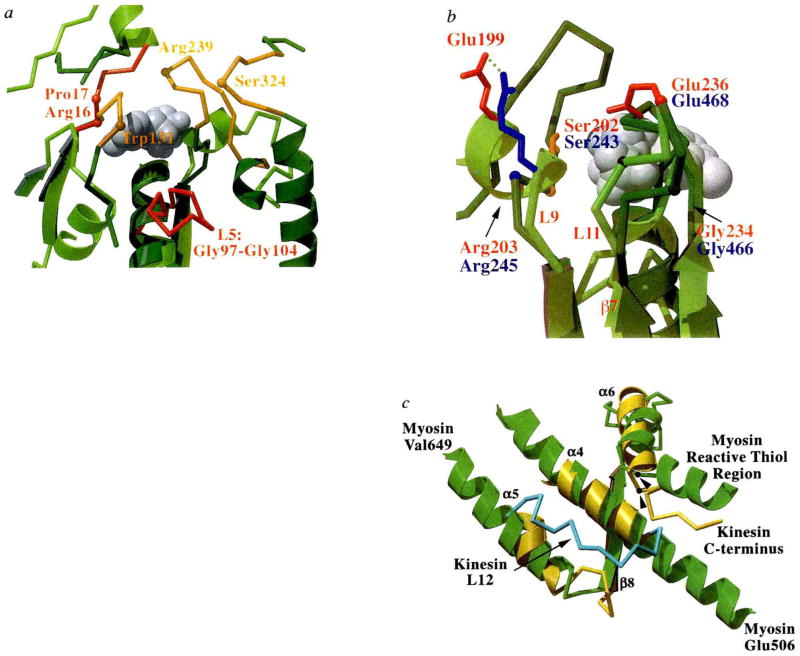
FIG. 3.
Nucleotide environment of kinesin and comparison of functional regions with myosin, a, Loops surrounding the entrance to the nucleotide-binding pocket are shown in this view of kinesin (light green with red loops) overlaid with myosin (dark green with orange loops). The bound ADP is shown as grey spheres, and highlighted residues and loops are discussed in the text. b, The back of the nucleotide-binding pocket with kinesin in light green and myosin in dark green. Residues that may be involved in γ-phosphate sensing are indicated (kinesin residues labelled in red, myosin in blue). Side chains are coloured red for Glu, blue for Arg, orange for Ser, and grey for Gly. Myosin residues are indicated by their α-carbon positions. The bound ADP is shown as grey spheres. This view has been rotated by approximately 180° relative to that in a. c, Structures potentially involved in transducing conformational changes from the nucleotide and filament binding site to a mechanical amplifier in kinesin (yellow) and myosin (green). Secondary structure elements of kinesin are indicated. Orientation is similar to that in Fig. 2b. The kinesin loop (L12; amino acids 272–280) that may be involved in microtubule binding is shown in light blue. The 142-amino-acid actin-binding domain of myosin is not shown, but is located between myosin residues Glu 506 and Val 649. Myosin’s reactive thiol region that may bend or melt during myosin’s ATPase cycle34 and the C terminus of the current kinesin model are indicated. The position of glycine residues that may be involved as pivot points for conformational movements are shown for myosin Gly 699 and kinesin Gly 319 (arrowheads).