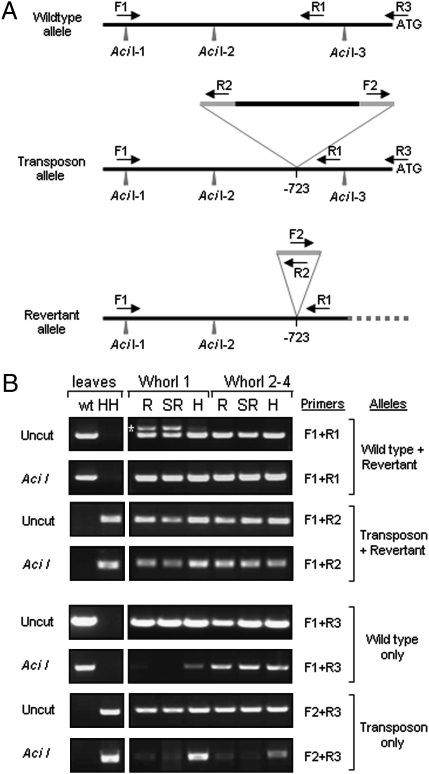
Fig. 4.
Retrotransposon excision is associated with demethylation of the PvGlo locus. (A) Organization of wild-type, transposon, and revertant alleles of PvGlo showing location of upstream methylation-sensitive AciI restriction enzyme sites. The PvGlo-specific primers (F1, R1, R3) and transposon-specific PCR primers (R2, F2) are indicated. The retrotransposon insertion site is shown (-723); LTRs are shown in gray. (B) Methylation-sensitive PCR analysis of the wild-type, transposon, and revertant alleles of PvGlo using allele-specific primer combinations as indicated. Primer specificity is demonstrated on DNA from leaves of wild-type (wt) and homozygous Hose in Hose (HH) plants. DNA samples from whorl-1 tissue or combined whorl-2, -3, and -4 tissue from revertant (R), semirevertant (SR), and Hose in Hose (H) flowers from the same plant were either digested with Aci1 or uncut as indicated before PCR amplification using allele-specific primer combinations as shown before fractionation by agarose gel electrophoresis. The different alleles amplified by each primer combination are indicated and the excision allele-specific PCR product is highlighted (*). PCR products generated from uncut genomic DNA that are absent after predigestion with AciI reveal unmethylated sites.