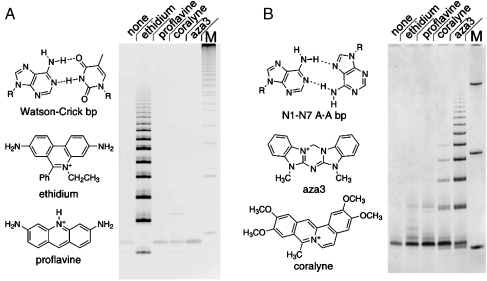
Fig. 5.
Base pair recognition by a given intercalator is a necessary, but not sufficient, condition for achieving intercalation-mediated ligation. (A) PAGE analysis of products from the condensation of d(pCGTA) (200 μM) in the absence and the presence of various small molecules (600 μM when present). Relative to the ligand-free reaction (Lane 1), ethidium (Lane 2) promotes the polymerization of d(pCGTA), whereas proflavine (Lane 3), another Watson–Crick intercalator, does not. Neither coralyne (Lane 4) nor aza3 (Lane 5), which binds homo-A duplexes, promotes d(pCGTA) ligation. (B) PAGE analysis of products formed by the condensation of d(pA6) (500 μM) in the absence and the presence of various small molecules (750 μM when present, lanes as in A). Relative to the ligand-free reaction (Lane 1), neither Watson–Crick intercalator promotes the ligation of d(pA6) (Lanes 2 and 3). In contrast, aza3 (Lane 5) promotes the ligation of d(pA6). Although coralyne also binds homo-A duplexes, it promotes the ligation of d(pA6) only weakly (Lane 4). The N1–N7 base pair shown for homo-adenine duplexes has been shown to be consistent with molecular modeling and chemical probing studies (33). Additional reaction details are provided in Materials and Methods. The gel Lane marked M in panel A contains marker bands generated by ligation of a tiling decanucleotide, the gel Lane marked M in panel B contains DNA oligonucleotides of length 10, 32, and 110 nt.