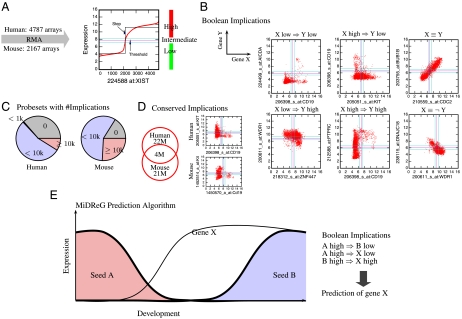
Fig. 1.
Computational prediction of developmental genes using Boolean implications. (A) BooleanNet algorithm on 4,787 Affymetrix U133 Plus 2.0 human microarrays and 2,167 Affymetrix 430 2.0 mouse arrays that were downloaded from NCBI’s Gene Expression Omnibus. (B) The scatter plots show six different types of Boolean implications between X and Y in human datasets. (C) The pie charts show the percentage of probesets with the indicated number of Boolean implications (0, < 1,000, < 10,000, and ≥10,000) in human and mouse datasets. More than 60% of the probesets have greater than 1,000 Boolean implications. (D) The Venn diagram shows the number of Boolean implications that are conserved across humans and mice. The mouse homologs were identified by using the euGene database: 15,199 human probesets and 10,695 mouse probesets have corresponding homologs. There are 4 M conserved Boolean implications out of 22 M in the human dataset. A conserved Boolean implication, KIT high⇒CD19 low is shown on the right. (E) MiDReG algorithm. It uses two seed genes: A, which is expressed early in development, and B, which is expressed later in the development, and identifies gene X by using Boolean implications, which is hypothesized to be expressed earlier than gene B and its expression is maintained throughout further development.