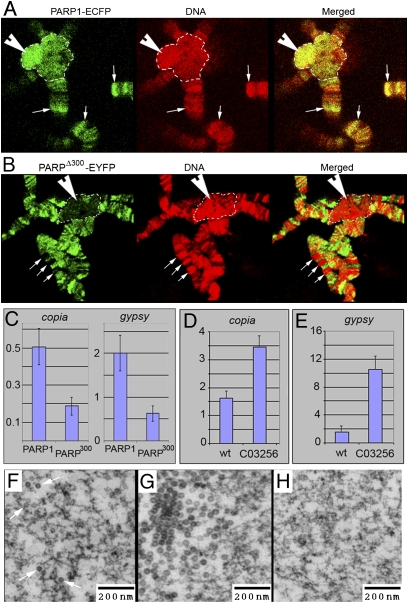
Fig. 5.
PARPΔ300 protein is excluded from the regions of heterochromatin. (A and B) Comparison of PARP1 and PARPΔ300 protein localization in salivary gland polytene chromosomes. Salivary glands were dissected from UAS::PARP1-ECFP, Arm::Gal4; ParpC03256/ParpC03256 (A) or UAS::PARPΔ300-EYFP, Arm::Gal4; ParpC03256/ParpC03256 (B) third-instar larvae, squashed on slides, and immunostained with anti-GFP antibody (green). DNA is shown in red. PARP1 protein shows significant accumulation in the regions of constitutive (arrowhead) and intercalary (arrows) heterochromatin (A), whereas PARPΔ300 is excluded from these regions (B). Constitutive heterochromatin is indicated with a dashed line. (C) ChIP assay demonstrates that chromatin of retrotransposons copia and gypsy accumulates significantly less PARPΔ300 protein than PARP1. (D–H) C03256 mutation disrupts silencing of retrotransposons. The quantitative RT-PCR assay using primers specific to copia (D) and gypsy (E) retrotransposons detects an elevated level of their RNA in ParpC03256 mutant larvae compared with heterozygous animals. EM microphotographs reveal the accumulation of retroviral particles in the nucleoplasm of ParpC03256 animals: individual particles (F) and clusters (G). (H) Silencing of retrotransposons could be restored by PARP1-DsRed expression.