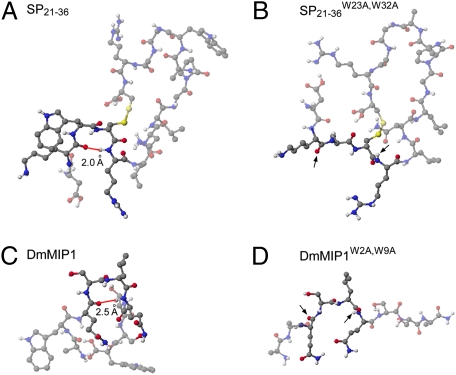
Fig. 3.
Conserved Trp residues stabilize beta-turn conformation in both SP and MIPs. Secondary structures of (A) SP21–36, (B) SP21–36W23A,W32A, (C) DmMIP1, and (D) DmMIP1W2A,W9A are predicted by low energy conformers calculated with MM2 and MOPAC (Materials and Methods). (A and C) In wild-type peptides, the distances between carbonyl groups of i amino acids and amino groups of I + 3 amino acids are determined in the range of hydrogen bonding distance (red line, 1.9–2.8 Å), a key signature of beta-turn conformation. (B and D) Trp > Ala mutations abolished the beta-turn conformations completely. The corresponding carbonyl groups of i amino acids and NH groups of I + 3 amino acids in mutants were indicated by arrows. Amino acids participating in the beta-turn conformations and their corresponding ones in mutants are highlighted. Black, blue, red, and white balls indicate carbon, nitrogen, oxygen, and hydrogen atoms respectively.