Abstract
This paper presents robust 3-D algorithms to segment vasculature that is imaged by labeling laminae, rather than the lumenal volume. The signal is weak, sparse, noisy, nonuniform, low-contrast, and exhibits gaps and spectral artifacts, so adaptive thresholding and Hessian filtering based methods are not effective. The structure deviates from a tubular geometry, so tracing algorithms are not effective. We propose a four step approach. The first step detects candidate voxels using a robust hypothesis test based on a model that assumes Poisson noise and locally planar geometry. The second step performs an adaptive region growth to extract weakly labeled and fine vessels while rejecting spectral artifacts. To enable interactive visualization and estimation of features such as statistical confidence, local curvature, local thickness, and local normal, we perform the third step. In the third step, we construct an accurate mesh representation using marching tetrahedra, volume-preserving smoothing, and adaptive decimation algorithms. To enable topological analysis and efficient validation, we describe a method to estimate vessel centerlines using a ray casting and vote accumulation algorithm which forms the final step of our algorithm. Our algorithm lends itself to parallel processing, and yielded an 8× speedup on a graphics processor (GPU). On synthetic data, our meshes had average error per face (EPF) values of (0.1–1.6) voxels per mesh face for peak signal-to-noise ratios from (110–28 dB). Separately, the error from decimating the mesh to less than 1% of its original size, the EPF was less than 1 voxel/face. When validated on real datasets, the average recall and precision values were found to be 94.66% and 94.84%, respectively.
Index Terms: Adaptive algorithms, GP-GPU, membrane segmentation, mesh generation, robust detection, vessel centerline extraction, vessel surface segmentation
I. Introduction
Accurate and rapid segmentation of microvasculature from 3-D images [1] is important for diverse studies in neuroscience [2], tumor biology [3], stem-cell niches [4], cancer stem cell niches [5], and other areas. It is needed for measuring vascular features such as surface areas, diameters, and tortuosities of vessel segments, branching patterns of the vascular tree [3], and distributions and orientations of cells relative to the vasculature [2]. When time-lapse imaging of living vasculature is performed, segmentation results can be used for analyzing angiogenesis [6]. Finally, quantifying the impact of pharmacological interventions requires vessel segmentation for change analysis [3], [6]. Vascular imaging methods can be classified into two categories from a molecular imaging standpoint. One approach is to use a contrast agent that labels the vessel lumen, by way of the blood flow [3]. Another is to use a contrast agent that labels the membranous vessel laminae. In the former case, vessels appear as a set of solid tubes that can be segmented using model-based segmentation algorithms [1]. In the latter case, the vessel structures appear as a set of hollow tubes in a nonisotropic space (imaging resolution is not the same for all dimensions), for which the prior literature on confocal image analysis does not contain methods for segmenting the laminae and vessel center-lines.
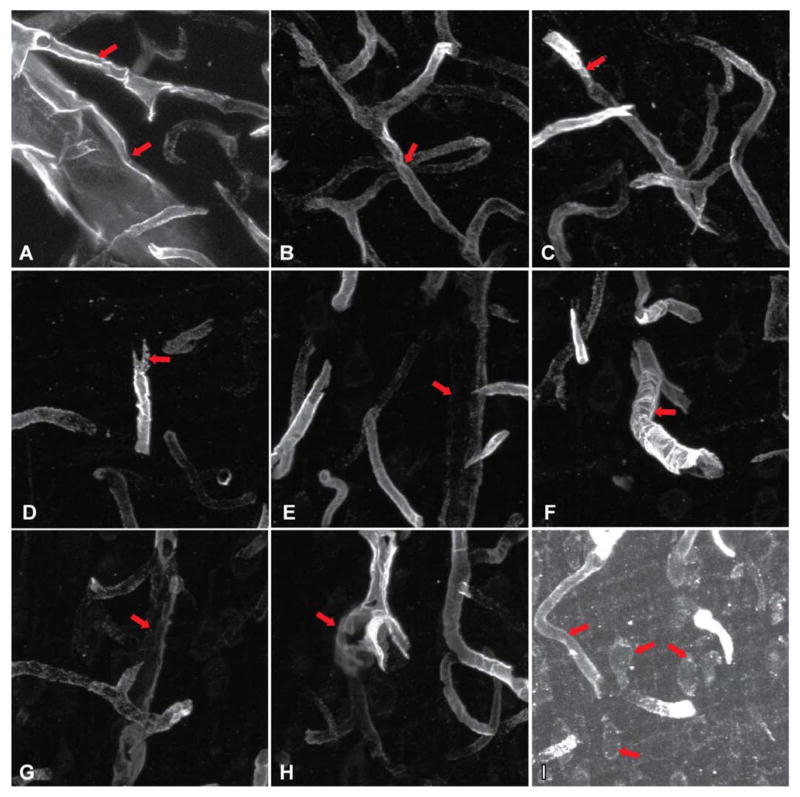
Fig. 1 illustrates the challenges. These images were produced by fluorescently tagging endothelial barrier antigen (EBA), and imaging its distribution using a laser-scanning confocal microscope. Even in projection images, it is clear that the vessels are far from ideal hollow tube-like structures. First, the specimen preparation steps deform the tissue. The inner membranes of vessels are thin, so the molar concentrations of the antigens bound by the contrast agents are low to begin with. The fluorescent signals are weak (usually quantum limited), and low-contrast, leading to a low signal-to-noise ratio. The fluorophore distribution is nonuniform, leading to gaps in the vessel surface, and varying contrast. With newly formed vessels of most interest, such as the results of tumor angiogenesis [3], and/or embryonic development [7], the vessel geometry is irregular and complex. In a wide field of view, one is also faced with vessels of widely varying diameters, and the finer vessels can be almost filled in terms of appearance. Finally, various types of imaging artifacts are found in images that must be rejected.
Fig. 1.
Gallery of sample images illustrating some of the challenges to surface segmentation. (a) An open vessel end with large variations in vessel diameters. (b) Closely situated vessel segments. (c) Abrupt variations in vessel signal. (d) Irregular broken vessel segments. (e), (g) Large signal variations and gaps. (f) Shows vessel with strong depth-dependent attenuation. (h) Low-contrast and blurry vessel signal with complex branching. (i) Contrast stretched image showing significant signal overlap with other fluorescent imaging channels (nuclear channel, in this case). Our segmentation algorithm has been designed to robustly detect the vasculature overcoming these issues.
This paper presents robust adaptive algorithms for computing a geometric mesh representation of the vessel surfaces and traces of their centerlines, together with estimates of the statistical detection confidence, local curvature, orientation (surface normal, inside/outside surfaces), thickness, and surface areas. Our method for adaptive vessel extraction takes advantage of the connectivity of the vessel laminae to intelligently reject imaging noise. We do not make assumptions regarding the geometry of the vessel laminae (e.g., cylindrical structure), so our method is also (in principle) applicable to segmentation of nonvessel membranes. Finally, our algorithms are amenable to parallel computation—we show that they can be implemented on graphics processing units (GPUs) commonly available on desktop computers.
A. Related Literature and Key Contributions
Four bodies of literature are relevant to this work: vascular segmentation algorithms, robust detection and estimation theory, mesh generation and 3-D visualization, and stream-oriented parallel computation. Automated vessel segmentation is a well-studied topic (e.g., see the review by Kirbas and Quek [1]). Methods for 3-D vessel segmentation have come a long way from the pioneering work by Verdonk et al. and others [8]–[11]. There is also an interest in segmenting nonvessel tubular structures such as neurites and cytoskeletal elements [12]–[15]. Broadly, algorithms can be classified into five major categories: 1) sequential tracing/vectorization [3], [12], [13], [16]–[18]; 2) matched filtering and vesselness based segmentation [19]–[21]; 3) skeletonization [14], [15]; 4) level sets and active contour based methods [22], [23]; and 5) graph-based surface reconstruction [24]–[27].
Sequential tracing algorithms [3], [16]–[18] work by following vascular segments starting from some initial seed points. Although some tracing algorithms are based on robust estimators [3], they produced unsatisfactory results for our images because they assume filled, rather than hollow tubes. As noted earlier, the literature on segmenting vessel laminae is sparse. We are aware of work on segmenting pulmonary tubes from lung CT imaging [28], but no methods for confocal microscopy. Matched filtering based approaches [20] model the vessel structure as the intensity-ridges of a multiscale vesselness function [20]. These methods are not applicable to hollow tubes. Also, they are susceptible to outliers and not robust to noise, as noted by Krissan et al. [20]. In principle, a Hessian filtering based plateness measure [29] could be used, but proved suboptimal in our experiments. Skeletonization based approaches are designed to seek out the medial axes of the tube-like structures from binarized/grayscale images [15]. Unfortunately, they do not produce the medial axes of hollow tubes. Active contour approaches can, in principle, adapt to various complex vessel intensity profiles, but suffer from the well-known “leakage” problem that is pronounced in our case. The “leakage” refers to the problem of active contours leaking into the background region from foreground in places of weak edges. This eventually continues to grow across a large background region until it meets another strong edge separating foreground and the background regions. Graph based algorithms seek an optimum cut in a weighted graph. The edge weights could be based on a similarity metric on the feature nodes [30] or on the likelihood of an edge at each voxel [26], [27]. Smoothness constraints are added to obtain a good reconstruction [24], [25]. Combining the 2-D segmentation of individual slices does not work well because the vessels are not oriented along any particular axis. The recent 3-D algorithm by Li et al. [25] uses vessel centerline information to unfold tubular objects. Li et al. do a surface segmentation of multiple coupled surfaces by computing a minimum closed set in a 4-D geometric graph. Although their results are impressive, their approach is not feasible in our case since it requires prior segmentation capable of extracting vessel centerlines. Li et al. use [28] to extract centerlines using a multiseed fuzzy-connectedness technique. Some graph cuts based approaches also require training [31].
Some earlier edge detection approaches involve smoothing and local hypothesis testing [32]–[34]. These approaches tend to be locally optimized and lack robustness. Weighted local variance (WLV) based edge detection was introduced by Law and Chung [35] for vessel boundary detection. This technique was shown to be robust enough to detect low contrast edges. This method does not produce the segmentation by itself but is used to drive active contour segmentation techniques to segment filled vessels in MRA images. Methods to apply robust detection and estimation methods to 2-D vessel segmentation have been described by Mahadevan et al. [21]. We have used similar α-ranked robust likelihood ratios to detect the vessel structures, with some differences. The present work is focused on detecting thin 3-D surfaces instead of local tube-like structures. It also differs in the way we model the intensity profiles—we model vessel laminae as locally-planar patches with intensity values exceeding the background level, so our work also extends to nonvessel laminae. We present an adaptive region growing algorithm that greatly improves segmentation performance for noisy data. Finally, we present a ray casting and voting based method to compute synthetic filled-tube representations that can be traced by existing algorithms. Related voting techniques exist in the literature [36]–[38]. For example, Parvin [36] use a voting scheme for segmenting cell nuclei.
Visualization of vasculature remains an important related topic [8]–[11]. Puig et al. [9] described various methods, e.g., maximum-intensity projection (MIP), and slice-to-slice composition. More recent methods render the triangulated mesh using simulated lighting, and colors to indicate local features. For visualizing raw image data, volume rendering algorithms are best [39], [40]. We use a combination of raw image volume rendering with surface rendering of the segmented output. To enable rapid rendering of large meshes, we have adopted decimation, and to improve the mesh quality we have used diagonal swapping methods similar to that of Cebral and Lohner [11]. Yim and Summers [10] discuss voxelation as an issue in visualization and use surface anti-aliasing to improve visual quality. Yim et al. [41] used a tubular coordinate system to generate the mesh that is intuitive for modeling tubular structures, but it starts from a manual/semiautomatic specification of the vessel axis which makes it infeasible in practice. A more recent paper by Huang et al. [42] addresses the issue of visualization by combined volume and surface rendering. Flores and Schmitt [43] use depth map/range map for visualizing segmented vessels. Depth mapping uses a ray casting approach, so it is slow for large datasets.
Given the data intensive nature of vessel surface segmentation, and the need for timely computation, it is valuable to consider the amenability of algorithms to parallel processing. An interesting opportunity is the use of low-cost graphics processors for general purpose scientific computing (GP-GPU [44], [45]). The recent development of C compilers and reusable libraries has simplified GP-GPU programming [45]. Our work contributes vessel surface segmentation algorithms to this emerging body of literature.
II. Proposed Method
A. Overview of the Method
The first step of our algorithm identifies high-confidence foreground voxels using a robust voxel-based generalized hypothesis test. The second step performs an adaptive region growing algorithm to identify additional foreground voxels while rejecting noise. This also yields relative estimates of detection confidence at each voxel. The third step uses the marching tetrahedra algorithm to link the detected foreground voxels to produce a triangulated 3-D mesh with watertight [46] isosurfaces. The mesh is smoothed using a volume-preserving algorithm to eliminate jagged facets, and adaptively decimated using an edge-collapsing and volume-preserving algorithm to produce the final mesh. Once this is complete, we estimate the local surface curvature at each triangle. Finally, we extract the vessel topology by generating a filled-in vessel that could be traced by tube tracing techniques to produce centerline estimates. The following sections explain the above steps in detail.
B. Robust Hypothesis Tests for Initial Detection of 3-D Surface Voxels
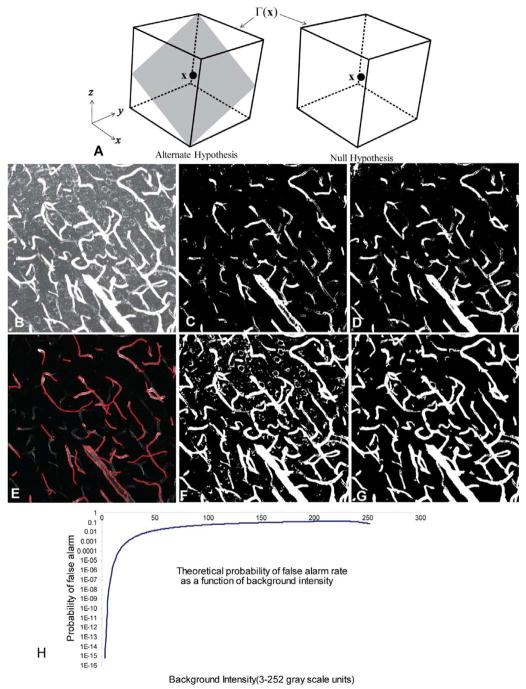
The image intensity is denoted s(x) where x = (x, y, z) is a 3-D voxel location. Let Γ(x) denote a rectangular 3-D neighborhood centered at x [Fig. 2(a)]. Let S(x) denote the vector of image intensity values in Γ(x). We define two hypotheses at each voxel, based on S(x). The null hypothesis (H0) is the case when the voxel belongs to the background of local intensity λb(x). The alternate hypothesis (H1) is the case when it belongs to the vessel surface with local foreground intensity λ1(x), and background intensity λ0(x). The vessel lamina is modeled as a planar surface of thickness 2ω within the neighborhood Γ(x) centered on voxel x. The 3-D angles of this plane are denoted Θ(x). Using the notation defined above, we define two conditional probability functions P[S(x)|H0, Γ, λb] and P[S(x)|H1, Γ, Θ, λ0, λ1], for the voxels in Γ(x), conditioned on the null and alternate hypotheses, respectively. Γ(x) is partitioned into two sets Γ1(x) and Γ0(x) denoting the foreground and background regions
Fig. 2.
Illustrating the performance of robust hypothesis testing based initial foreground detection compared to other approaches. (a) In the alternate hypothesis, there is a locally planar patch of the vessel surface passing through x within the rectangular neighborhood Γ(x). In the null case, all of the voxels in the neighborhood belong to the background. (b) maximum-intensity projection of the original 3-D confocal dataset that is contrast stretched to better show the imaging noise and overlap from the nuclear channel (small circular objects). (c) Adaptive Otsu thresholding. (d) Hessian filtering followed by adaptive Otsu thresholding. (e) Direct tube tracing results [4]. (f) Basic surface extraction algorithm described in Section II-B. (g) Results of adaptive region growth (Section II-D). These two panels have the same sensitivity (true positive rate). (h) The variation of the false alarm rate (with median estimate) for various background intensity values (5×5×5 neighborhood), for the case when λT = 3.
We assume a Poisson model for P[.] since weak fluorescence data are known to be quantum limited [47]. Hypothesis H0 is characterized by parameter λb, and H1 is characterized by parameters λ0 and λ1. Under the approximation that the voxel intensities within the neighborhood are mutually independent, the generalized likelihood ratio test to identify surface voxels is given by [48]
| (1) |
where τ is the threshold (1 in our case). The λ̂s and Θ̂ are the optimum values computed over all possible (λ, Θ) values.
Several authors have noted [49]–[51] that the ratio test in (1) is not robust to outliers. A single “bad” intensity value can un-acceptably change the estimated values of λ̂’s and the overall likelihood ratio. To overcome this problem, we use the robust generalized log-likelihood ratio of rank-ordered intensity values given by [51]
| (2) |
where η(x) is either 0 or 1. It is set to 0 whenever the voxel intensity s(x) lies in the highest or lowest α percentile of intensity values in both the background and foreground regions of the neighborhood Γ(x). Using this methodology, robust estimates of the parameters can be formulated as
| (3) |
To carry out this optimization, we discretize the 3-D angles Θ(x) to N equally-spaced values, where N is a function of the neighborhood size—larger neighborhoods can accommodate a finer discretization, and vice versa. Our algorithm iterates over these discrete values. For each value of the angle, we define the following three sets of voxels:
| (4) |
where the parameters (a, b, c) define the plane for a chosen value of Θ. The constraint |ax + by + cz| < ω represents a thick planar region of width 2ω. Using this notation, the parameters λ̂1, λ̂2, & λ̂b are estimated from the inliers as follows:
| (5) |
where |.| denotes the cardinality of a set. Without additional constraints, the alternate hypothesis will always fit the data better than the null hypothesis because it has one extra parameter. Since we expect the foreground and background regions to have a contrast between them, we constrain the minimum contrast expected in these regions. In effect, we do an optimization of (3) with constraint λ̂1(x) − λ̂0(x) ≥ λ̂T. Voxels with robust log-likelihood ratio ≥ 0 are considered foreground.
We consider the choice of α next. Note that as α approaches the 50th percentile, the α-ranked mean approaches the median estimate. The median estimator offers the highest-possible breakdown value. As an added advantage, it is possible to derive exact analytical formulae for the theoretical false-alarm rates (Section II-C). Computing these for a general α value would require numerical computation. Other α values may be justified in certain applications depending upon the nature of the imaging artifacts. If the noise field has a low tail, then lower values of α are preferable.
C. Impact of Varying the Contrast Parameter
We analyze the impact of varying the contrast parameter λ̂T for the most common case, when α = 50%, and the noise model is Poisson [47]. For simplicity, suppose that the neighborhood Γ(x) contains an odd number of voxels (2n + 1). We assume that the voxel intensity values are modeled by a probability mass function f and the corresponding cumulative distribution function is denoted F. The median value of (2n + 1) Poisson-distributed samples with mean λ is distributed according to the probability mass function gn,λ(m) given by (see the Appendix for more details and proof)
| (6) |
where
| (7) |
Let Γ1(Θ̂), Γ0(Θ̂) denote the foreground and background regions corresponding to the optimum value Θ̂. We compute the distribution of the median estimates (u, v) of the foreground and background regions as PU(u, Θ̂) ~ gΓ1(Θ̂),n1,λ 1 and PV(v, Θ̂) ~ gΓ0(Θ̂),n0,λ 0 as given by (6) and(7). Γ1(Θ̂), Γ0(Θ̂) subscripts are used to distinguish intensities coming from the foreground and background regions. The probability mass function for the distribution of w1 = u − v can be identified numerically since we have a modest number of discrete values (256 for 8-bit image data). The result is given below for the 8-bit case
| (8) |
The probability of a Type II error (missing a foreground voxel) is given by
| (9) |
Similarly, let w0 = p − q for the background intensity parameters, where PP ~ gn1,λb and PQ ~ gn01, λb
We find fw0 in a similar way to find the False alarm probability (Type I error) given by
| (10) |
Although the above estimates are computed numerically, they are exact since the domain is discrete. Fig. 2(h) shows a plot of the probability of false alarm as a function of the background intensity value λb, when λT is held constant at 3. From this we see that for any background intensity less than 50, the probability of false alarm will be less than 1% when λT = 3. In our real data samples, the background noise’s gray scale intensity varies between 0 and 30, with an average of 7. Therefore, our estimator is highly specific. Higher values of λT result in even smaller false alarm rates, making the test more specific, but less sensitive. To quantify the segmentation confidence, we use the robust log-likelihood log LR(S; Θ, λ1, λ0, λb) in (2). Higher values of log-likelihood indicate greater confidence that a voxel is indeed a surface voxel. The best corresponding λ̂1(x) − λ̂0(x) value is also a measure of the local contrast between the foreground and the background, and is another measure of confidence. We map this onto the segmented results as a color encoding (Fig. 3).
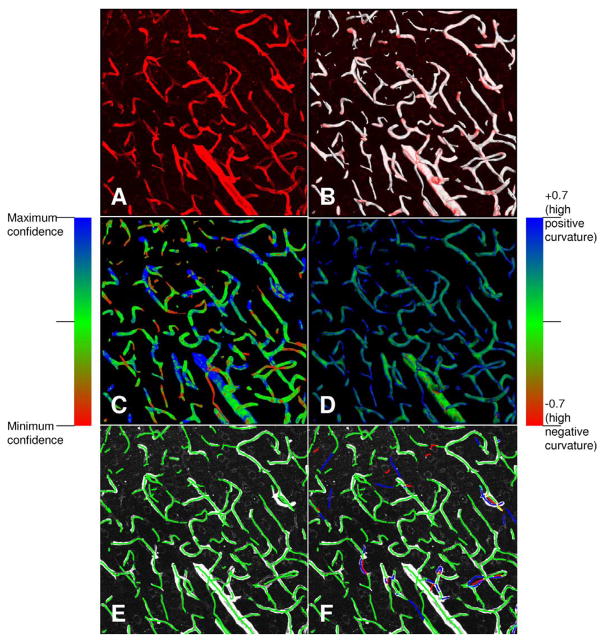
Fig. 3.
Illustrating the automated vessel segmentation results for a confocal image from the cortical region of the rat brain (field size: 368.55 μm × 368.55 μm × 93.00 μm with field resolution 0.36 μm × 0.36 μm × 1.5 μm giving rise to 1024 × 1024 × 63 voxels). (a) Maximum-intensity projection of the grayscale image data displayed in shades of red. (b) Automated segmentation results rendered as a surface (white) viewed axially. The grayscale data is overlaid on the rendering to enable simultaneous viewing of the raw data and the segmentation. (c) Display of the segmentation confidence estimates over the 3-D field using the indicated color map. (d) Display of the local curvature estimates using the indicated color map. (e) Result of automatic vessel centerline extraction. (f) Illustrating manual editing of centerlines by an expert observer for edit-based validation.
D. Adaptive Region Growth of the Initial Surface Voxel Set
The algorithm described in the previous section is effective at detecting foreground voxels, but is limited by the fact that the decision at each voxel is based solely on local intensity information within Γ(x). The sensitivity and specificity of the hypothesis tests conducted at each voxel are ultimately limited by the choice of the contrast parameter λT. At high sensitivity, this produces thick surfaces and isolated “noisy” background points in the segmentation. At low sensitivity, biologically interesting microvascular endings are missed. In order to achieve a better tradeoff, we present an adaptive region growing algorithm that takes advantage of the known continuity of the vasculature. In essence, we initially segment the prominent (high confidence) vessel regions and adaptively “grow” the segmentation to add new contiguous points from the lower-contrast regions (representing finer microvasculature). This has the important advantage of picking up the dimmer vessel points while intelligently rejecting background noise.
The initial segmentation is conducted with a high value of λT, denoted . The second phase of the algorithm uses the initial segmentation results as seed points and conducts a breadth-first search for additional contiguous surface points, driven by the segmentation confidence values described above. For every point in the 26-connected neighborhood of seed points that is not already a surface voxel, the algorithm checks if its contrast value (difference in estimated λ values) is within a difference of k to that of the current voxel and greater than . The value of k is set empirically (≈ 8). Higher values lead to thicker vessels, while a very low value leads to broken/undetected segments. Since the number of neighbors to be searched for each voxel is bounded by 26, the time complexity of the algorithm is proportional to the number of voxels in the image. This step is thus very fast. Importantly, it inherently rejects the isolated faint blob-like spectral imaging artifacts representing nuclei from another imaging channel, since they are not connected to the initial segmentation, even as it picks up the fainter small-bore vessels that may have a comparable intensity to these artifacts. This algorithm can be thought of as region growing, but with the addition of initialization and region growth decision making mechanisms that are appropriate for hollow vessel segmentation.
Overall, the choice of the lower threshold is much more important than . The latter is only used for initial seed generation and subsequent threshold values for the nearby voxels must be close to the actual threshold obtained from the data and are therefore independent of the used. We only have to make sure that the initial seed segmentation picks up at least one point in each disconnected vessel segment. On the other hand, a good choice of is necessary to avoid picking up spurious vessels from the data. A pseudo-code description of the basic-algorithm is presented in the Appendix and the adaptive region growing algorithm is presented in the supplement.
E. Automated Mesh Generation and Optimization
Hypothesis testing yields a binary-valued grid of surface points, but not a connected surface. For this, we generate a mesh representation of the vessel laminae that offers multiple advantages. At the simplest level, a mesh provides a major data reduction, so interactive visualization of a mesh is still much faster than ray-casting based volume rendering. We can leverage efficient geometric algorithms for smoothing, resampling, rescaling, and estimating features such as normal vectors (required for extracting vessel centerlines), curvature and vessel thickness from a mesh representation. It is straightforward to display features such as curvature and confidence values by way of color coding on the mesh representation. Finally, it is straightforward to overlay surfaces over volume renderings of raw voxel data to enable validation and editing. The segmentation is converted to a 3-D “watertight” (in the sense defined by [46]) mesh by generating the isosurface. For this, we use the “marching tetrahedra” algorithm [52]–[54] that addresses some ambiguities in the classical marching cubes algorithm [55]. In order to form a connected mesh, we maintain a hashing container that hashes the 3-D coordinate of a vertex to a pointer corresponding to the vertex element in the half-edge [56] mesh data structure. This has been done to avoid duplication of vertices and produce a connected oriented mesh. The resulting isosurface follows the points closely, but could be jagged-looking. While jaggedness can be removed by smoothing, one has to make sure that the volume does not shrink in the process. Another opportunity is to reduce the mesh complexity by decimation to accelerate visualization, and scale-specific estimation of features such as surface area. Besides, since we are not seeking subvoxel resolution in our segmentation, we do not require an unnecessarily dense mesh.
Laplacian mesh smoothing is commonly used since it is fast. However, it shrinks convex surfaces and expands concave ones, and alters the object volume. While this does not alter the overall topology, and thus may be useful for modeling coarsely sampled data, it does not do a good job when distances of a few voxels matter [4], [57]. To overcome this limitation, we use the volume-preserving smoothing algorithm of Kuprat et al. [58] in which edges are moved close to the center of the neighboring vertices while preserving the enclosed volume. This algorithm is slower and takes hundreds of iterations to get a smooth tube. In our work, we limit the number of iterations to about 10–20 as a tradeoff between speed and smoothness.
In order to visualize large datasets efficiently, and to estimate scale-specific features like curvature robustly, we reduce the complexity of the smoothed mesh by smoothing and iteratively collapsing the mesh edges. The literature has several papers on mesh decimation and related optimizations [58], [59]. Boundary preservation is not an issue in our work because we have a watertight mesh. To address the issue of volume preservation, we use the volume preserving edge collapsing algorithm described by Lindstrom and Turk [59]. Edge collapsing involves removal of two vertices and the edge between them, and replacing them with a single vertex. The triangles connected to these vertices must be reconnected consistently. We also perform edge swapping to improve the mesh’s appearance as we iteratively decimate, using the method described by Freitag et al. [60]. Edge swapping involves swapping the diagonals of a plane quadrilateral to make the mesh more symmetric and less skewed. As in the case of mesh smoothing, the volume preservation requirement restricts the new vertex to a plane of points. So, we add more constraints to uniquely identify the new position. We identify the point closest to the midpoint of the two vertices being removed as the new location for the vertex. We do not use priority queues of edge lengths for determining the order of edge collapses. Collapsing an edge involves change of edge lengths of all the connected edges. Therefore, the time complexity for the update of priority queue is high [61], [62]. We thus use a threshold on the edge lengths instead. The threshold is iteratively increased when there is no edge left to be collapsed. We also keep a threshold on the confidence of the triangle to be decimated. This is also iteratively reduced, to facilitate decimation of high-confidence regions before low-confidence fine structures. This allows us to adaptively decimate the mesh. Typically we decimate the mesh to 15% of its original size. The run time of this step is about a minute for a mesh size of 1.5 million triangles. In Section III, we show that this does not lead to a significant error (< 0.5 voxels per face).
After generating the mesh, we compute features such as surface normals, local surface curvature, and vessel wall thickness (where it exists) at each triangle of the mesh. Surface normals are readily available from the marching tetrahedra algorithm. Estimating local surface curvature requires a more careful estimation since it is both direction-dependent and sensitive to noise. For this, we use the algorithm described by Sacchi et al. [63]. We used this algorithm to estimate the curvature along the direction of maximum curvature, but averaged it over a line of eight triangles along the direction of maximum curvature to make it robust to local noise. To estimate the local vessel wall thickness, we cast a ray from each triangle opposite to its normal direction and compute the distance to the first intersecting face.
F. Vessel Centerline Extraction by Ray Casting and Vote Accumulation
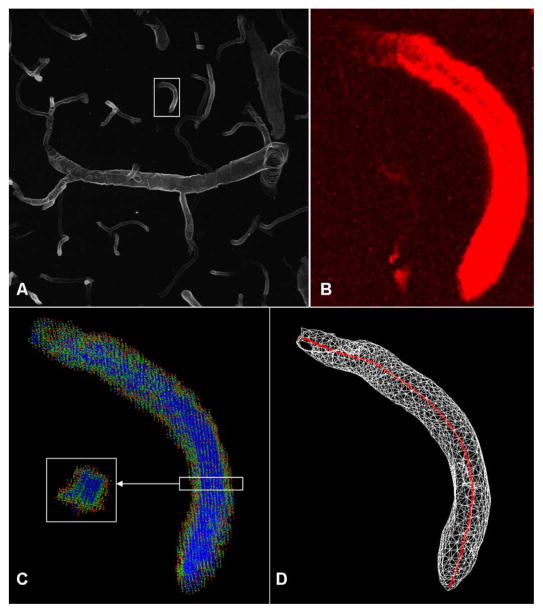
The segmented mesh described in the previous section is useful in its own right, and also provides the basis for estimating vessel centerlines. An important issue is the presence of gaps in the vessel surfaces. To estimate centerlines in a manner that is robust to gaps, we propose an approach based on ray casting and vote accumulation. Rays are cast uniformly in all directions from all points of the image volume (or a subsampled subset) to the segmented mesh. The projections of these rays cast on the normal of the first triangle encountered are computed and summed. We refer to this as “votes” accumulated by the point. We limit the maximum length of the rays to the maximum expected vessel radius. Points perfectly inside the cylindrical vessel accumulate the highest votes since large numbers of rays align with the normal vectors. Points outside the vessel accumulate the fewest votes since only a small portion of the cast rays meet the segmented vessels and even then not all of them are aligned with the normal vector of the surface triangle (Fig. 4). As we approach the interior boundary of the vessel, the normal vectors are less aligned with the cast rays and the vote accumulation decreases. This approach works even when the vessel boundary is incomplete. As an aside, note that this procedure would work better for spherical geometries than cylindrical, but the fluorescence imaging yields a pure image with mainly vessels. The vote accumulation image can be traced using tube tracing algorithms. In our examples, we used the algorithm of Tyrrell et al. [3]. These centerlines provide topological information, and can be edited much more efficiently compared to meshes (to correct errors, and for edit-based validation).
Fig. 4.
Illustrating the voting based centerline extraction method. (a) maximum-intensity projection of a confocal dataset. (b) Small vessel segment from panel A shown enlarged and volume rendered. (c) Voting results overlaid on the surface rendering, using the same color map as in Fig. 3. Small sliced region shows the cross-section. (d) Extracted centerline shown in red along with the surface rendering.
G. Parallel Implementation on Graphics Processors
Our algorithms are suited to parallel computation. Graphics processing units (GPUs) of desktop computers have emerged as a powerful and accessible form of parallel computing. The nVIDIA 8800GTX (G80) processor used by us has 128 32-bit processors organized as a bank of 16 streaming multiprocessors (SM), each containing eight streaming processors and two super function units (SFU), all organized as a single instruction multiple data (SIMD) stream processor. A stream is a set of records requiring similar computation. The GPU runs a single kernel at a time. A kernel is a function that is applied in parallel to multiple records of the stream. This function has some restrictions—for each element one can only read from the input, perform operations on it, and write to the output. It is permissible to have multiple inputs and multiple outputs, but never a section of memory that is both readable and writable. Each SM has 8 K registers, 16 KB of shared cache memory, and 64 KB of constant memory that cannot be modified by kernel functions, but can be written from the host computer. An important advance represented by the G80 is the general-purpose (nongraphics) programming model named CUDA (Compute Unified Device Architecture) that is simpler compared to traditional graphics programming. In this model, the GPU is viewed as co-processor to the host CPU with its own DRAM (device/global memory). The computation is structured as a set of “blocks” that run in parallel. Each block consists of multiple “threads.” Data-parallel portions of an application execute on the device as kernels that run many cooperative threads in parallel. There is no cooperation between blocks.
Notwithstanding its relative simplicity, CUDA is still a relatively unsophisticated parallel programming model. It presents some challenges when implementing 3-D surface segmentation compared to traditional parallel multiple instructions multiple data (MIMD) architectures. For example, memory locking mechanisms to prevent concurrent updates to the same memory location cannot be taken for granted. Also, multiprocessors in CUDA work in a SIMD fashion, where best performance is achieved when all core processors perform the same operation. Divergence among core processors in the same multiprocessor results in serialization of the different paths taken, and causes a performance penalty. The GPU’s on-chip cache memory is shared by all threads of a block and not across blocks. The latency for read/write in shared memory is about 100–150 times lesser than device memory which makes it desirable for repeatedly used data [64].
We focused on parallelizing two data-intensive parts of the algorithm. First, we consider the processing associated with a single voxel (Section II-B), and a single seed point for the adaptive region growth (Section II-D). Next, we consider the processing of multiple voxels/seed points simultaneously. The steps are illustrated in Fig. 5(c). A cube of voxels centered on a voxel being tested is loaded in parallel into low-latency shared memory from high latency device memory by a block of threads. The number of voxels surrounding the voxel depends on the window size parameter. The load instructions are uniformly divided between the threads. The parameters needed for the hypothesis testing are loaded into the shared memory before processing begins. To calculate the medians of the foreground and background regions, the neighborhood voxels are sorted. An efficient CPU implementation of sorting is “quicksort” with complexity O(n log n). Since CUDA does not support recursive functions, quicksort is not appropriate. Hence, we adopted the parallel bitonic sort instead [65]. A block of threads perform parallel bitonic sorting to sort the cube of voxels in the shared memory. Bitonic sorting can be implemented more efficiently if the number of threads matches the number of data points to be sorted. In our case, this is not always true because the GPU requires the number of threads to be a power of 2 [64]. To handle this, we sorted the array in multiple passes. Each time we sort as many elements as the number of threads available. For example, when we have 256 threads and 343 elements to be sorted, we first sort the first 256 elements, then the last 256 elements and finally the first 256 elements [Fig. 5(b)]. In order for this method to always sort correctly, there is a constraint on α and β which turns out to be α < β/3 [α, β defined in Fig. 5(b)]. This constraint holds true in our case. Also, we sort both the foreground and background in the same array because loading the foreground and background elements in different arrays would lead to parallel queue loading problem and has to be serialized. We merely negate all the background elements and offset by −1 to make sure their range is nonoverlapping with the foreground range. We then sort the array using the above-mentioned method and find the corresponding medians.
Fig. 5.
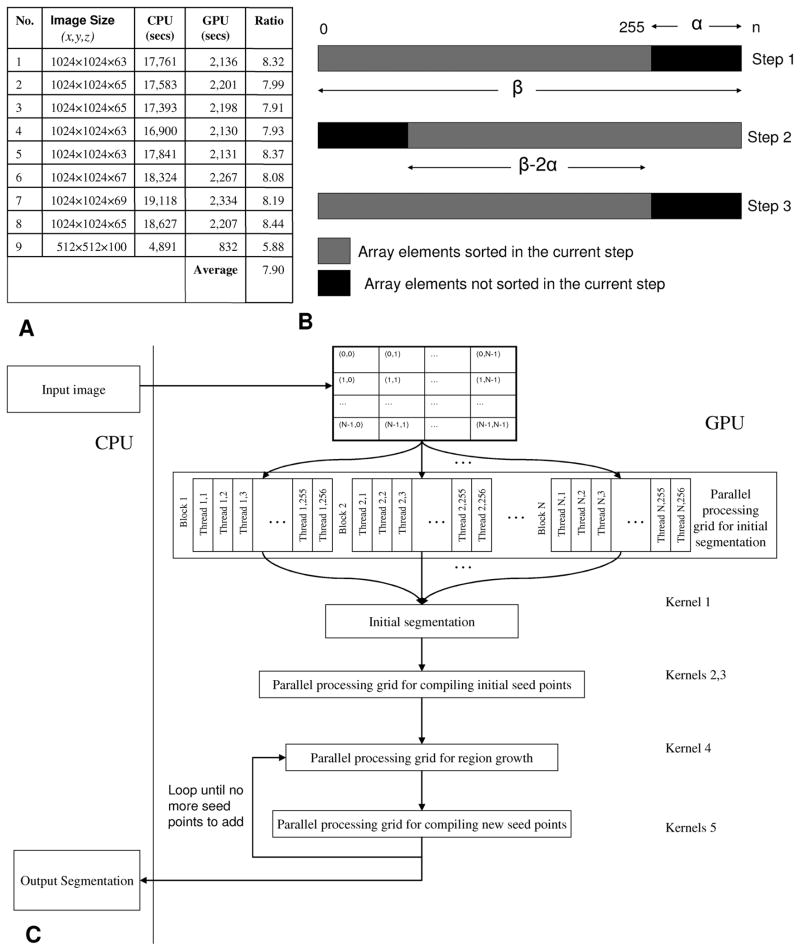
Illustrating the parallel implementation in GPU. (a) Table of runtimes of the CPU code and the GPU code. (b) Illustration of the multipass bitonic sorting used in the GPU implementation. (c) Flowchart of the surface segmentation algorithm with adaptive region growing step. We implemented five different kernel functions to achieve a parallel implementation in the GPU.
Our CPU implementation used breadth-first search based on a queue data structure to accomplish the seed growth. But queues cannot be implemented efficiently on GPU [44]. So, a parallel breadth-first search was implemented on the GPU: each thread was made to process one seed point. This involves the 26 neighbors of the seed point that must be analyzed and marked as the next level seed points under the region growing conditions of the adaptive algorithm (Section II-D). Since the number of seed points varies dynamically, we vary the number of blocks of threads dynamically by choosing the optimal number of blocks for each case. Hussein et al. have shown that this can be implemented efficiently on bounded degree graphs [44]. We maintained a 2-D Boolean matrix which is has as many rows as the number of current seed points and 26 columns. One kernel initially identifies all the new points to be grown and populates this matrix. Then, we identify the total amount of memory to be allocated for the new seed points and allocate them. We used the parallel prefix scan for parallel counting and computing the total number of seed points [66]. Another kernel identifies all the non-repeating x, y, z coordinates and makes a list of them (because the size of the list is already known, this can be done without any divergence of threads). This process is repeated until we have no more seed points to process.
III. Experimental Results
A. Experiments With Phantom Data
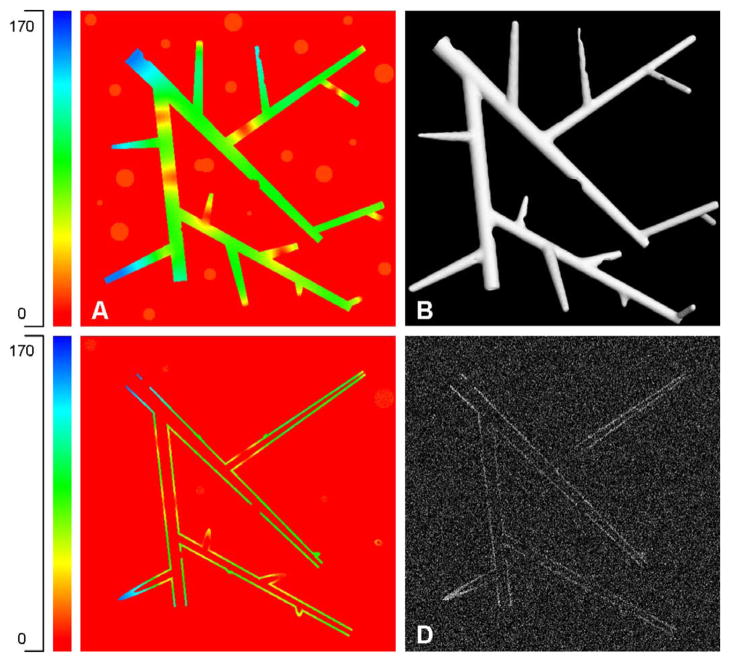
To characterize the performance of the proposed algorithms, we computationally synthesized the 3-D phantom image shown in Fig. 6 incorporating common simulated defects found in real images, including slow fading of vessel signal, missing signal, varying radii, branching, cross-channel interference in multi-spectral imaging systems. The structural and intensity parameters used to generate the phantom images were realistic. The phantom includes eight points of abrupt cuts in the vessel signal, 15 points of fading, and 19 points of cross-channel interference introduced by adding spheres of low intensity (to simulate spectral unmixing artifacts). The intensity of the spheres was made equal to the points of low intensity in the fading parts of vessel signal to evaluate the effectiveness of the adaptive region growing algorithm. The phantom image also includes regions where the vessel surface smudges to become a filled vessel of very low radius. This is observed in real fluorescence images, particularly in regions of low intensity.
Fig. 6.
Showing the 3-D vessel phantom (512 × 512 × 100 × 8-bit) used for our experiments. (a) Maximum-intensity projection displayed using the colormap (grayscale units) shown on the left. The phantom was designed to simulate varying diameters, intensity values, and the presence of spectral unmixing artifacts (faint blobs in the background). (b) 3-D surface rendering of the phantom’s vessel structure. (c) Slice 19 of the phantom displayed using the colorscale on the left to show the hollow nature of the vessels. (d) Slice 19 data shown with added noise (Gaussian, σ = 90).
We computed synthetic data using 2 imaging models: Poisson, and additive i.i.d. Gaussian noise, Since, the background intensity is low compared to the foreground, the Poisson sampling does not fully allow us to evaluate the robustness of the algorithm at all signal-to-noise ratios. For this, we also generated datasets with additive Gaussian noise, even though the algorithm is based on Poisson modeling. The mesh generated from the ground truth vessel, denoted MP, is taken as the ground truth mesh for evaluating segmentation results. Specifically, if MS denotes the mesh for a segmentation algorithm to be evaluated, we define a performance metric termed the “error per face (EPF)” based on the minimum 3-D distance dmin(.) between each triangle in MS, denoted tS, to the nearest triangle of MP, and the vice-versa. These two distance values were averaged to compute the EPF measure, as follows:
| (11) |
This metric (expressed in units of voxels per face) punishes both false positives and false negatives. In case of a high false positive rate, the average error from every face of the noisy image’s mesh to the nearest face of ground truth mesh would be high, and in case of high false negative rate the average error from every face of the ground truth mesh to nearest face of noisy image’s mesh would be high. The averaged EPF values were plotted against the averaged peak signal-to-noise ratio (PSNR) value of the noisy image. The PSNR between two equal-sized 8-bit grayscale images (A, B) is given by [67]
| (12) |
where
| (13) |
where N is the number of voxels in the images. A high value of PSNR indicates a low difference between the two images while a low value of PSNR indicates a large difference. We use the PSNR values for evaluating both Poisson and Gaussian noise models.
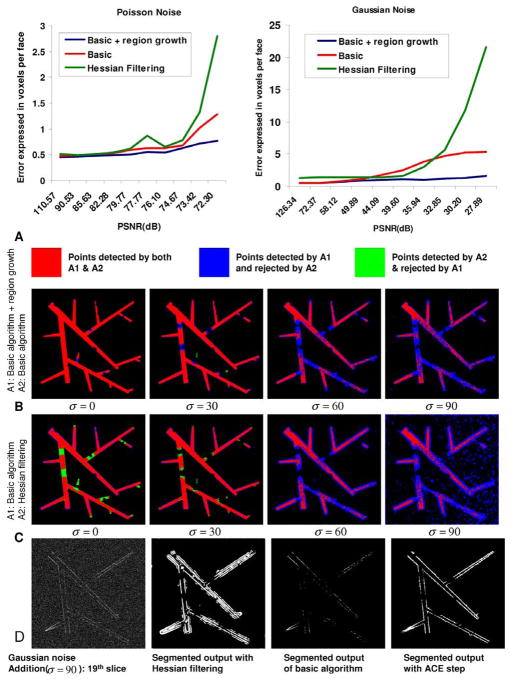
Simulated images corrupted by Poisson noise were computed by varying the background intensity in the range from 5 to 45, in steps of five gray scale units. We compared the performance of the algorithm, with, and without the adaptive region growing step. Table I shows the segmentation parameters used for segmenting the phantom images. Fig. 7(a) shows the resulting plot of EPF as a function of the PSNR. Overall, the proposed algorithm exceeds the performance of Hessian filtering based technique, and produces adequately accurate results in the presence of Poisson noise (EPF measure ranging from 0.45–0.77 voxel per face for PSNR values in the range 110–72) even without the adaptive region growth. Adaptive region growth always improves the performance. To evaluate the algorithm at lower PSNR values, we added Gaussian noise. The standard deviation of the Gaussian noise σ was varied in the range 0–90 grayscale units to achieve a PSNR range of 126–28. In this realm, the adaptive region growth results in a much greater performance improvement. Specifically, the EPF values for the basic algorithm ranged from a fraction of a voxel per face all the way to 5.4 voxels per face. In comparison, the EPF for the same datasets with the adaptive region growth stays below 1.6 voxel per face.
TABLE I.
Parameter Settings for Segmentation of Phantom Images
| Poisson Noise Model | Gaussian Noise Model | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Background Intensity | λT |
|
|
Γ | Standard Deviation | λT |
|
|
Γ | ||||
| 0 | 12 | 20 | 3 | 5×5×5 | 0 | 12 | 12 | 12 | 5×5×5 | ||||
| 5 | 12 | 20 | 5 | 5×5×5 | 10 | 15 | 45 | 15 | 5×5×5 | ||||
| 10 | 12 | 20 | 5 | 5×5×5 | 20 | 18 | 50 | 15 | 5×5×5 | ||||
| 15 | 12 | 20 | 7 | 5×5×5 | 30 | 35 | 55 | 18 | 7×7×7 | ||||
| 20 | 14 | 20 | 7 | 5×5×5 | 40 | 45 | 55 | 20 | 7×7×7 | ||||
| 25 | 16 | 20 | 10 | 5×5×5 | 50 | 55 | 60 | 20 | 7×7×7 | ||||
| 30 | 16 | 20 | 10 | 5×5×5 | 60 | 65 | 60 | 25 | 7×7×7 | ||||
| 35 | 16 | 20 | 10 | 5×5×5 | 70 | 70 | 60 | 30 | 7×7×7 | ||||
| 40 | 18 | 20 | 10 | 5×5×5 | 80 | 75 | 60 | 30 | 7×7×7 | ||||
| 45 | 18 | 20 | 10 | 5×5×5 | 95 | 75 | 65 | 30 | 7×7×7 | ||||
Fig. 7.
Results on phantom image data. (a) Shows a plot of the average error per face (EPF) measure as a function of the peak signal-to-noise ratio (PSNR) for the case of Poisson and additive white Gaussian noise respectively for three algorithms. (b) Compares the best performance of basic algorithm with/without region growth in the presence of Gaussian noise for various values of the noise standard deviation (σ). The adaptive region growing step clearly picks up more vessel content without picking up the noise. (c) Compares the performance of the basic algorithm over the Hessian filtering based algorithm. (d) Shows one single slice of the noisy image data for Gaussian noise with σ = 90 along with the segmentation results from the Hessian filtering, basic algorithm, and adaptive region growth, respectively.
To provide the reader with a visual feel for the results, the row in Fig. 7(b) shows a comparison of the basic algorithm against the algorithm with the adaptive region growth for the case when the Gaussian noise model is used, and σ = 0, 30, 60, 90, respectively. The results are presented as 2-D projections of 3-D results. They are color coded as follows: red indicates voxels that were detected by both algorithms being compared; blue indicates voxels that were picked up by the first algorithm, but missed by the second; green indicates voxels that were picked up by the second algorithm, but missed by the first; and black indicates voxels that were missed by both algorithms. At lower noise level, the performance of the basic algorithm and adaptive region growth are comparable. At the higher noise level (σ = 90), the relative improvement provided by the region growth is abundantly obvious. In order to provide the reader with a widely-known benchmark for the basic algorithm (without region growth) we compared it against the Hessian based plateness filter [29] followed by adaptive Otsu thresholding [68], as shown in Fig. 7(c). At lower noise level, the Hessian filtering picks up more vessel content than basic algorithm, but produces thicker vessels than the ground truth throughout the image. The performance of the Hessian filtering degrades significantly at higher noise levels. The Hessian plateness function was computed based on the eigenvalues of the Hessian matrix. The “plateness” function was computed based on the “vessel-ness” measure suggested in the paper with a minor change to detect plates instead of vessels. We used the ratio of λ1/λ3 and λ2/λ3 for ℜa and ℜb, respectively. Note, the λs here denote the Eigen values of the Hessian matrix computed in [29]. ℜa and ℜb refer to the geometric ratios based on a second-order ellipsoid used in computing the plateness measure [29]. The first term was a simple Gaussian instead of an inverted Gaussian. The value of a, b parameters [29] were fixed at 0.5. The value of c was fixed at one half of the maximum Hessian norm. The range of scale value σ was optimized individually for each noise level. Since computing a Hessian image is very memory intensive, we also broke down the image into 4 parts each with about 20 pixels wide extra border and computed the Hessian individually and merged the fragments. To provide the reader with another view, Fig. 7(d) shows a sample slice (#19) of the 3-D image for the case when σ = 90. Again, the proposed algorithm with adaptive region growing step outperforms others.
We measured the performance of the decimation by iteratively decimating the mesh for the phantom (MP). Even after reducing the mesh to less than 1% of its original size, the EPF remained less than 1 voxel per face. Decimation achieves a tradeoff between error in the surface and complexity of the mesh representation. In summary, our Phantom based experiments clearly demonstrate the validity of the proposed algorithms, and the value of adaptive region growing algorithm. Accordingly, our experiments with real data always included the region growing step.
B. Real Data Experiments
Confocal images were collected from the rat brain cortex. The Wadsworth Center Institutional Animal Care and Use Committee (IACUC) approved all animal procedures. Adult male Sprague-Dawley rats were anesthetized with a ketamine/xylazine mixture, and transcardially perfused with 200 mL warm (37 °C) phosphate buffered saline (PBS) followed by 200 mL 4% paraformaldehyde in PBS using a constant-pressure system [69]. Brains were removed and immersion fixed in 4% paraformaldehyde for an additional 24 h, then washed in HEPES-buffered Hanks’ saline (HBHS) containing azide (90 mg/L). Horizontal 100-μm-thick tissue slices were cut using a vibratome (FHC, Inc.). Histochemistry was performed as follows. Tissue slices were incubated in 5 mg/mL NaBH4, washed in HBHS, incubated in 5% BSA in HBHS, and then incubated overnight in primary antibody identifying endothelial cells (mouse anti-EBA, Sternberger Monoclonals, Inc., Lutherville, MA). After washing, sections were incubated overnight in secondary antibody (Alexa 405 anti-mouse, Invitrogen, Eugene, OR). Following washing, sections were mounted in ProLong Gold (Invitrogen). Spectral imaging was performed on a Zeiss LSM 510 META system using a 405 nm laser and a 25 × W/0.8 NA. apochromat objective. Images were 1024 × 1024 pixels. Voxel resolution for all images was 0.36 μm × 0.36 μm × 1.5 μm along the (X, Y, Z) axes, respectively.
Fig. 2 shows the effectiveness of the technique. An adaptive Otsu algorithm with a conservative 128 × 128 × 63 window yields a partial segmentation of the vasculature [Fig. 2(c)]. Hessian filtering followed by adaptive Otsu thresholding yields a similar result [Fig. 2(d)]. A recent tube tracing algorithm [3] that is designed for tracing vessel lumens produces poor traces [Fig. 2(e)]. The robust hypothesis test described above extracts a much more complete segmentation [Fig. 2(f) and (g)]. Fig. 2(g) shows the value of the adaptive region growing step in comparison with the basic detection algorithm [Fig. 2(f)]. It clearly rejects most of the blob-like artifacts from the nuclei channel interference since it takes advantage of the connectivity of vasculature. Note, Fig. 2(g) and (f) are computed for the same true-positive rate for a fair comparison.
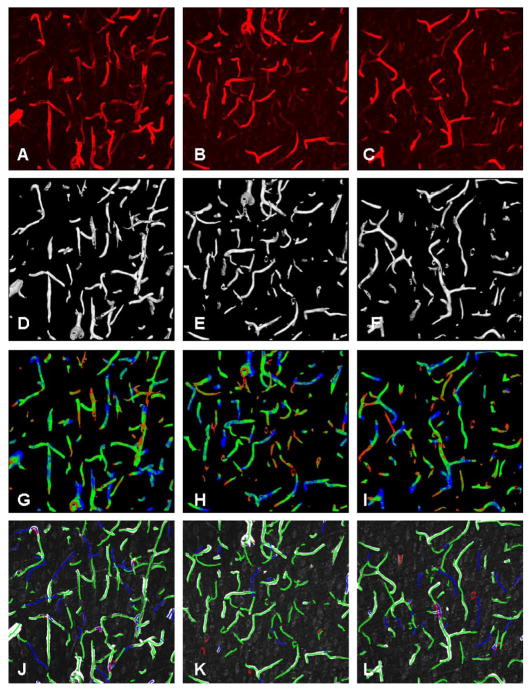
Fig. 3 shows the results of automated segmentation of a confocal dataset. In Fig. 3(b), we overlaid the grayscale data on the mesh. In Fig. 3(c), we overlaid the confidence values. In Fig. 3(d), we overlaid the curvature map. Fig. 3(e) shows the result of automated centerline extraction and Fig. 3(f) shows the result of manual edit-based validation of the centerlines. The idea behind edit-based validation is for a human expert to inspect the automated results and make corrective edits that are recorded and analyzed. There are two important outcomes from edit-based validation: 1) centerline extraction results that are fully acceptable to the biological expert and 2) performance data for the automated algorithms. Fig. 8 shows segmentation results for three representative datasets (more examples are in the supplement). The topmost row in these figures shows a volume rendering of the original image data. The next row shows a smoothed mesh representing the automatic segmentation (with region growing step), using a decimation factor of 4. The third row shows the same mesh as in the middle column, but with a color map indicating the confidence values. The color map is shown in Fig. 3. Overall, the proposed algorithm is robust and effective in extracting the visible vascular structure in diverse confocal datasets studied by us to date. All the images were 60–70 slices of size 1024 × 1024 pixels. They were automatically segmented with , N = 21 and a 5 × 5 × 5 neighborhood using adaptive region growth.
Fig. 8.
Segmentation results on real data. (a)–(c) Volume rendering of the image data. (d)–(f) 3-D rendering of the mesh generated from the segmentation. (g)–(i) Confidence map of the segmentation is mapped over the segmentation (color scale shown in Fig. 6). (j)–(l) Results of edit-based validation (green lines are computed centerlines, blue lines were added by the human expert, and red lines were deleted).
To quantify the centerline extraction performance on real datasets, an expert neuroscientist manually validated six 3-D datasets using an edit-based strategy [3]. The domain expert is provided with a graphical editing tool to add, delete or modify parts of centerlines. For this, the raw image intensities were amplified 5 times to reveal faint vessels during the validation. The last row of Fig. 8 shows the results in the form of color-coded centerlines. Green lines indicate valid automatically traced centerlines. Manually added segments are marked in blue and the deleted segments are marked in red. The vast majority of these added centerlines were very low contrast and noisy segments with contrast comparable to that of the nuclear channel interference signal that can be inferred with difficulty by an expert observer. Recovering these segments automatically would require higher-order constraints and perceptual organization principles [38]. The lengths of the edited centerlines were recorded. From these annotations, the average recall and precision (defined in [3]) were found to be 94.66% and 94.84%, respectively. The average recall and precision values were based on the lengths of correctly identified centerlines.
C. Parallel Programming Results
We chose representative commercial products from both of the GPU and CPU markets: an NVIDIA Geforce 8800 GTX which has a core clock speed of 575 MHz, a memory size of 768 MB with 86.4 GB/s memory bandwidth. For the CPU timings, we used a traditional dual-core AMD Opteron 244(1.81 GHz with 3 GB RAM). The GPU code was developed using NVIDIA’s CUDA API. The CPU code was compiled under Visual Studio 7 with optimization level O2 and SSE2. The CPU implementation was single threaded. The GPU results were compared against the CPU results under Windows XP. For the same input data, the speed was calculated by comparing the wall-clock time required by the CPU and GPU. Times are measured after the file I/O, but do include PCI-E bus transfer times. The panel A of Fig. 5 shows timing results for nine sample datasets. Overall, efficient memory usage and parallelizing techniques have reduced the total processing time of the algorithm by roughly 8 ×. The bulk of the computing time (72%) is taken up by the data-intensive first step. In this step, we perform hypothesis testing for all points and store them before we do adaptive region growing. The remaining kernels use 15% of the runtime. Copying data back and forth between the GPU and the main memory uses 13% of runtime.
IV Conclusion
The software and supplements are available from the authors’ Web site1. This work provides an effective solution to the problem of accurately segmenting vessel surfaces, and vessel centerlines from molecular images that label laminae rather than the lumenal flow [3]. Our methods are also applicable to nonvascular membranes, but an actual demonstration is deferred to future work. The need for this type of algorithm is widespread due to the pervasive importance of vasculature. For example, investigations in tumor biology require vessel segmentation for quantifying angiogenesis, evaluating drug candidates, and mapping the locations and movements of various cells (e.g., pericytes) relative to the vasculature. In stem-cell science, it is important to quantify the perivascular distribution of progenitors and other cells in the stem-cell niche. Cancer stem cells too have a perivascular distribution not unlike stem cells [5]. In the area of brain mapping, Bjornsson et al. [2] have demonstrated cytovascular mapping of brain tissue for brain studies.
The design principles used in this work, especially the use of robust statistical methods and adaptive region growing are broadly applicable. The resulting algorithm is capable of handling a wide range of PSNR values. Our algorithm outperforms the Hessian based plateness filters, in spite of simplifying voxel independence assumptions. Although our algorithm is fully automated, there remain a few user-settable parameters that depend on the noise level of the data set. In future work, automated parameterization could eliminate the need to initialize parameters for every data set [70]. Finally, there is a need for effective graphical tools for editing remaining errors [71].
Even serial implementations of our method are practical. Beyond this, the practicality of a parallel implementation on low-cost GPUs is gratifying, and sets the stage for high-throughput studies. Further speedups are possible using page-locked host memory, texture memory access, asynchronous memory copies overlapped with arithmetic computations and atomic functions. Keeping in mind that GPUs increase in performance faster than Moore’s law, future speedups will occur without reprogramming
Acknowledgments
The computational portions of this research were supported by the Center for Subsurface Sensing and Imaging Systems, under the Engineering Research Centers Program of the National Science Foundation (Award Number EEC-9986821), and by NIH/NIBIB under Grant R01 EB005157. The neuro-science research that led to this work was supported by grants from the NIH, NIBIB, EB-P41-002030(WS), NINDS, NS-R01-044287(WS), and NS051531 (BR).
The authors would like to thank V. Mahadevan for helpful discussions.
Appendix
Algorithm 1: Basic Vessel Segmentation Algorithm
| S ← Read Image |
| for every x ∈ domain(S) do |
| Γ ← [x − w, x + w] × [y − w, y + w] × [z − w, z + w] |
| Compute η(x) |
| Compute optimum λb, for the given η |
| Lmax ← −1 |
| for i = 1 to N do |
| Compute optimum λ0, λ1 for the given η |
| if then |
| Continue |
| else |
| Compute the α-ranked likelihood ratio L |
| if Lmax ← L then |
| posmax ← i |
| Lmax ← L |
| λC = λ1 − λ0 |
| end if |
| end if |
| end for |
| if Lmax > 0 then |
| if then |
| Mark(x) ← true |
| end if |
| Normal(x) ← posmax |
| Likelihood(x) ← Lmax |
| contrast(x) ← λC |
| end if |
| end for |
Proof for the Expression for Median Distribution of a Finite Set
Claim
The median value of 2n + 1 Poisson-distributed samples with mean λ is distributed according to the probability mass function gn,λ (m) given by
| (14) |
where
| (15) |
Proof
Assuming that the 2n + 1 voxel intensity values are organized as a linear array, we estimate the probability that the median of these values is some value m. For this, the element at the nth position in this array must be m. The median distribution for the continuous case is given by [72], [73]
| (16) |
where x is a continuous variable representing and idealized notation for the voxel intensity value. The main difference between median distributions in a discrete case to that of a continuous case arises in the case when the median value is repeated. Suppose we have r repetitions of the median value m in the last (n + 1) values, the probability of this event is given by
| (17) |
where Pn,r(m) is the probability of the first (n + r) elements being less than or equal to m with at least last r elements being equal to m. This is easy to see because the probability of the last (n + 1 − r) elements being greater than m is given by (1 − F(m))n+1− r and there are possible choices. Summing the probability in (17) over the possible values of r yields (14).
To understand (15), consider the first n + r elements. Let the last r + k elements be equal to m and the rest less than m (where k greater than zero ensures that at least r elements be m). The probability of the first n − k elements being less than m is given by F(m − 1)n−k and the probability of r + k elements to be equal to m is given by f(m)r+k. The number of ways of choosing the r + k elements from n + r elements is given by . Summing over all valid values of k yields (15). Q.E.D.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Arunachalam Narayanaswamy, Email: naraya3@rpi.edu, Department of Electrical, Computer and Systems Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180 USA.
Saritha Dwarakapuram, Email: saritha.dwarakapuram@am.sony.com, Department of Electrical, Computer and Systems Engineering, Rensselaer Polytechnic Institute, Troy 12180 NY. She is now with the U.S. Research Center, Sony Electronics, Inc., San Jose, CA 95131 USA.
Christopher S. Bjornsson, Email: bjornc@rpi.edu, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180 USA
Barbara M. Cutler, Email: cutler@cs.rpi.edu, Department of Computer Science, Rensselaer Polytechnic Institute, Troy, NY 12180 USA
William Shain, Email: wshain@mac.com, Center for Neural Communication Technology, Wadsworth Center, New York State Department of Health, Albany, NY 12201 USA.
Badrinath Roysam, Email: roysam@ecse.rpi.edu, Department of Electrical, Computer and Systems Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180 USA.
References
- 1.Kirbas C, Quek FKH. A review of vessel extraction techniques and algorithms. ACM Comput Surv. 2004;36:81–121. [Google Scholar]
- 2.Bjornsson CS, Lin G, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B. Associative image analysis: A method for automated quantification of 3d multi-parameter images of brain tissue. J Neurosci Meth. 2008;170(1):165–78. doi: 10.1016/j.jneumeth.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyrrell JA, Tomaso ED, Fuja D, Tong R, Kozak K, Jain RK, Roysam B. Robust 3-D modeling of vasculature imagery using superellipsoids. IEEE Trans Med Imag. 2007 Feb;26(2):223–237. doi: 10.1109/TMI.2006.889722. [DOI] [PubMed] [Google Scholar]
- 4.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311(5769):1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese C, Poppleton H, Kocak M, Hogg T, Fuller C, Hamner B, Oh E, Gaber M, Finklestein D, Allen M. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Tyrrell JA, Mahadevan V, Tong RT, Brown EB, Jain RK, Roysam B. A 2-D/3-D model-based method to quantify the complexity of microvasculature imaged by in vivo multiphoton microscopy. Microvasc Res. 2005;70(3):165–178. doi: 10.1016/j.mvr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Liebling M, Forouhar AS, Wolleschensky R, Zimmermann B, Ankerhold R, Fraser SE, Gharib M, Dickinson ME. Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development. Develop Dynamics. 2006;235(11):2940–2948. doi: 10.1002/dvdy.20926. [DOI] [PubMed] [Google Scholar]
- 8.Verdonck B, Bloch L, Maitre H, Vandermeulen D, Suetens P, Marchal G. Accurate segmentation of blood vessels from 3D medical images. Proc. Int. Conf. Image Process; Lausanne, Switzerland. 1996. pp. 311–314. [Google Scholar]
- 9.Puig A, Tost D, Navazo I. An interactive cerebral blood vessel exploration system. Proc. 8th Conf. Visualizat; Phoenix, AZ. Oct. 1997; pp. 443–446. [Google Scholar]
- 10.Yim PJ, Summers RM. Analytic surface reconstruction by local threshold estimation in the case of simple intensity contrasts. Proc. SPIE Med. Imag. 1999: Physiol. Function Multidimensional Images; 1999. pp. 288–300. [Google Scholar]
- 11.Cebral JR, Lohner R. From medical images to CFD meshes. Proc. 8th Int. Meshing Roundtable; South Lake Tahoe, CA. Oct. 1999; pp. 321–331. [Google Scholar]
- 12.Al-Kofahi KA, Lasek S, Szarowski DH, Pace CJ, Nagy G, Turner JN, Roysam B. Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Trans Inform Tech Biomed. 2002 Jun;6(2):171–187. doi: 10.1109/titb.2002.1006304. [DOI] [PubMed] [Google Scholar]
- 13.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58A(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 14.Koh IYY, Lindquist WB, Zito K, Nimchinsky EA, Svoboda K. An image analysis algorithm for dendritic spines. Neural Comput. 2002;14(6):1283–1310. doi: 10.1162/089976602753712945. [DOI] [PubMed] [Google Scholar]
- 15.He W, Hamilton TA, Cohen AR, Holmes TJ, Pace CJ, Szarowski DH, Turner JN, Roysam B. Automated three-dimensional tracing of neurons in confocal and brightfield images. Microsc Microanal. 2003;9(4):296–310. doi: 10.1017/S143192760303040X. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Zhang H, Li H, Hu G. An improved algorithm for vessel centerline tracking in coronary angiograms. Comput Methods Programs Biomed. 2007;88(2):131–143. doi: 10.1016/j.cmpb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Hart M, Holley L. A method of automated coronary artery tracking in unsubtracted angiograms. Proc. Comput. Cardiol; 1997. pp. 93–96. [Google Scholar]
- 18.Park S, Lee J, Koo J, Kwon O, Hong S. Adaptive tracking algorithm based on direction field using ML estimation in angiogram. Proc. IEEE TENCON; Brisbane, Australia. 1997. pp. 671–675. [Google Scholar]
- 19.Gang L, Chutatape O, Krishnan SM. Detection and measurement of retinal vessels in fundus images using amplitude modified second-order Gaussian filter. IEEE Trans Biomed Eng. 2002 Feb;49(2):168–172. doi: 10.1109/10.979356. [DOI] [PubMed] [Google Scholar]
- 20.Krissian K, Malandain G, Vaillant R, Trousset Y, Ayache N. Model-based multiscale detection and reconstruction of 3-D vessels. Proc. CVPR; Santa Barbara, CA. 1998. pp. 722–727. [Google Scholar]
- 21.Mahadevan V, Narasimha-Iyer H, Roysam B, Tanenbaum HL. Robust model-based vasculature detection in noisy biomedical images. IEEE Trans Inform Tech Biomed. 2004 Sep;8(3):360–376. doi: 10.1109/titb.2004.834410. [DOI] [PubMed] [Google Scholar]
- 22.Holtzman-Gazit M, Goldsher D, Kimmel R. Hierarchical segmentation of thin structures in volumetric medical images. Proc. MICCAI; Montreal, QC, Canada. 2003. pp. 562–569. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Amini AA. Quantifying 3-D vascular structures in MRA images using hybrid PDE and geometric deformable models. IEEE Trans Med Imag. 2004 Oct;23(10):1251–1262. doi: 10.1109/TMI.2004.834612. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Holzapfel G, Schulze-Bauer C, Stollberger R, Thedens D, Bolinger L, Stolpen A, Sonka M. Segmentation of wall and plaque in vitro vascular MR images. Int J Cardiovasc Imag. 2003;19(5):419–428. doi: 10.1023/a:1025829232098. [DOI] [PubMed] [Google Scholar]
- 25.Li K, Wu X, Chen DZ, Sonka M. Optimal surface segmentation in volumetric images—A graph-theoretic approach. IEEE Pattern Anal Mach Intell. 2006 Jan;28(1):119–134. doi: 10.1109/TPAMI.2006.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thedens DR, Skorton DJ, Fleagle SR. Methods of graph searching for border detection in image sequences with applications to cardiac magnetic resonance imaging. IEEE Trans Med Imag. 1995 Mar;14(1):42–55. doi: 10.1109/42.370401. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Chen DZ. Optimal net surface problems with applications. Proc. 29th Int. Colloq. Automata, Languages, Programm; 2002. pp. 1029–1042. [Google Scholar]
- 28.Tschirren J, Hoffman EA, McLennan G, Sonka M. Intrathoracic airway trees: Segmentation and airway morphology analysis from low-dose CT scans. IEEE Trans Med Imag. 2005 Dec;24(12):1529–1539. doi: 10.1109/TMI.2005.857654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. presented at the MICCAI; Cambridge, MA. 1998. [Google Scholar]
- 30.Malik J, Shi J. Normalized cuts and image segmentation. IEEE Pattern Anal Mach Intell. 2000 Aug;22(8):888–905. [Google Scholar]
- 31.Boykov YY, Jolly M-P. Interactive graph cuts for optimal boundary and region segmentation of objects in N-D images. Proc Int. Conf. Comput. Vis; 2001. pp. 105–112. [Google Scholar]
- 32.Qiu P, Bhandarkar SM. An edge detection technique using local smoothing and statistical hypothesis testing. Pattern Recognit Lett. 1996;17(8):849–872. [Google Scholar]
- 33.Mascarenhas NDA, Prado LOC. Edge detection in images: A hypothesis testing approach. Proc. Int. Joint Conf. Pattern Recognit; 1978. pp. 707–709. [Google Scholar]
- 34.Newsam GN. Edge and line detection as exercises in hypothesis testing. Proc. Int. Conf. Image Process; 2004. pp. 2685–2688. [Google Scholar]
- 35.Law MWK, Chung ACS. Weighted local variance-based edge detection and its application to vascular segmentation in magnetic resonance angiography. IEEE Trans Med Imag. 2007 Sep;26(9):1224–1241. doi: 10.1109/TMI.2007.903231. [DOI] [PubMed] [Google Scholar]
- 36.Parvin B, Yang Q, Han J, Chang H, Rydbery B, Barcellos MH. Iterative voting for inference of structural saliency and characterization of subcellular events. IEEE Trans Image Process. 2007 Mar;16(3):615–623. doi: 10.1109/tip.2007.891154. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, Parvin B. Harmonic cut and regularized centroid transform for localization of subcellular structures. IEEE Trans Biomed Eng. 2003 Apr;50(4):469–476. doi: 10.1109/TBME.2003.809493. [DOI] [PubMed] [Google Scholar]
- 38.Medioni G, Lee M, Tang C. A Computational Framework for Segmentation and Grouping. 1. New York: Elsevier; 2000. [Google Scholar]
- 39.Cabral B, Cam N, Foran J. Accelerated volume rendering and tomographic reconstruction using texture mapping hardware. Proc. Symp. Visualizat; 1994. pp. 91–98. [Google Scholar]
- 40.Higuera FV, Hastreiter P, Naraghi R, Fahlbusch R, Greiner G. Smooth volume rendering of labeled medical data on consumer graphics hardware. Proc. SPIE Med. Imag. 2005: Visualizat., Image-Guided Procedures, Display; Apr. 2005; pp. 13–21. [Google Scholar]
- 41.Yim PJ, Cebral JJ, Mullick R, Marcos HB, Choyke PL. Vessel surface reconstruction with a tubular deformable model. IEEE Trans Med Imag. 2001 Dec;20(12):1411–1421. doi: 10.1109/42.974935. [DOI] [PubMed] [Google Scholar]
- 42.Huang A, Nielson GM, Razdan A, Farin GE, Baluch DP, Capco DG. Thin structure segmentation and visualization in three-dimensional biomedical images: A shape-based approach. IEEE Trans Visual Comput Graph. 2006 Jan;12(1):93–102. doi: 10.1109/TVCG.2006.15. [DOI] [PubMed] [Google Scholar]
- 43.Flores JM, Schmitt F. Segmentation, reconstruction and visualization of the pulmonary artery and the pulmonary vein from anatomical images of the visible human project. Proc. 6th Mexican Int. Conf. Comput. Sci; Sep. 2005; pp. 136–144. [Google Scholar]
- 44.Hussein M, Varshney A, Davis L. On implementing graph cuts on CUDA. presented at the 1st Workshop General Purpose Process. Graphics Process. Units (GPGPU); Boston, MA. 2007. [Google Scholar]
- 45.Buck I. GPU computing with NVIDIA CUDA. ACM SIGGRAPH 2007 Courses; Aug. 2007. [Google Scholar]
- 46.Dey TK, Goswami S. A water-tight surface reconstructor. J Comput Inf Sci Eng. 2003 Dec;3(4):302–307. [Google Scholar]
- 47.Pawley JB. Handbook of Confocal Microscopy. 2. New York: Plenum; 1995. [Google Scholar]
- 48.Besag J. On statistical analysis of dirty pictures. J R Statist Soc B. 1986;48(3):259–302. [Google Scholar]
- 49.Huber PJ. A robust version of the probability ratio test. Ann Math Statist. 1965;36(6):1753–1758. [Google Scholar]
- 50.Poor HV. An Introduction to Signal Detection and Estimation. 2. New York: Springer-Verlag; 1994. [Google Scholar]
- 51.Huber PJ. Robust Statistics. New York: Wiley; 1981. [Google Scholar]
- 52.Chan SL, Purisima EO. A new tetrahedral tesselation scheme for isosurface generation. Comput Graph. 1998;22(1):83–90. [Google Scholar]
- 53.Lorensen WE, Cline HE. Marching cubes: A high resolution 3D surface construction algorithm. Proc. SIGGRAPH; 1997. pp. 163–170. [Google Scholar]
- 54.Bloomenthal J. Graphics Gems. IV.8. New York: Academic; An implicit surface polygonizer; pp. 324–349. [Google Scholar]
- 55.Matveyev SV. Approximation of isosurface in the marching cube: Ambiguity problem. Proc. IEEE Conf. Visualizat; Washington, DC. 1994. pp. 288–292. [Google Scholar]
- 56.Guibas L, Stolfi J. Primitives for the manipulation of general subdivisions and the computation of Voronoi diagrams. ACM Trans Graphics. 1985;4(2):74–123. [Google Scholar]
- 57.Sun Y, Goderie S, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45(6):873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 58.Kuprat A, Khamayseh A, George D, Larkey L. Volume conserving smoothing for piecewise linear curves, surfaces, and triple lines. J Comput Phys. 2001;172(1):99–118. [Google Scholar]
- 59.Lindstrom P, Turk G. Fast and memory efficient polygonal simplification. Proc. IEEE Conf. Visualizat; Research Triangle Park, NC. 1998. pp. 279–286. [Google Scholar]
- 60.Freitag LA, Gooch CO. Tetrahedral mesh improvement using swapping and smoothing. Int J Numer Methods Eng. 1997;40(21):3979–4002. [Google Scholar]
- 61.Hoppe H. Progressive meshes. Proc. SIGGRAPH; 1996. pp. 99–108. [Google Scholar]
- 62.Cutler B, Dorsey J, McMillan L. Simplification and improvement of tetrahedral models for simulation. Proc. Eurographics Symp. Geometry Process; 2004. pp. 93–102. [Google Scholar]
- 63.Sacchi R, Poliakoff JF, Thomas PD, Hafele K-H. Curvature estimation for segmentation of triangulated surfaces. Proc. 2nd Int. Conf. 3DIM; Ottawa, ON, Canada. 1999. pp. 536–543. [Google Scholar]
- 64.NVIDIA CUDA Programming Guide. NVIDIA Corp; Santa Clara, CA: 2007. [Google Scholar]
- 65.Ionescu MF. Optimizing parallel bitonic sort. Proc. 11th Int. Parallel Process. Symp; 1997. pp. 303–309. [Google Scholar]
- 66.Sengupta S, Harris M, Zhang Y, Owens JD. Scan primitives for GPU computing. Proc. Graphics Hardware; 2007; San Diego, CA. 2007. pp. 97–106. [Google Scholar]
- 67.Natarajan B, Bhaskaran V, Konstantinides K. Low-complexity block-based motion estimation via one-bit transforms. IEEE Trans Circuits Syst Video Technol. 1997 Aug;7(4):702–706. [Google Scholar]
- 68.Jain R, Kasturi R, Schunck BG. Machine Vision. New York: McGraw-Hill; 1995. [Google Scholar]
- 69.Olsen KR. Preparation of fish tissues for electron microscopy. J Electron Microsc Tech. 1985;2(3):217–228. [Google Scholar]
- 70.Abdul-Karim MA, Roysam B, Dowell-Mesfin NM, Jeromin A, Yuksel M, Kalyanaraman S. Automatic selection of parameters for vessel/neurite segmentation algorithms. IEEE Trans Image Process. 2005 Sep;14(9):1338–1350. doi: 10.1109/tip.2005.852462. [DOI] [PubMed] [Google Scholar]
- 71.Zorin D, Schröder P, Sweldens W. Interactive multiresolution mesh editing. Proc. 24th Annu. Conf. Comput. Graphics Interactive Tech; 1997. pp. 259–268. [Google Scholar]
- 72.Chu JT. On the distribution of the sample median. Ann Math Statist. 1955;26(1):112–116. [Google Scholar]
- 73.Rider PR. Variance of the median of samples from a Cauchy distribution. J Amer Statist Assoc. 1960;55(290):322–323. [Google Scholar]