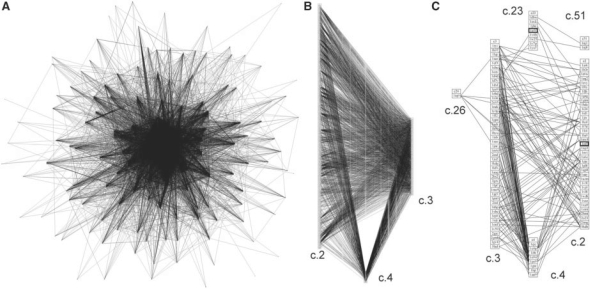
Fig. 1.
Protein structure similarity networks for α/β proteins, classical Rossmannoids and a subset of motifs. (A) Edges were drawn by connecting representatives of each fold when the two motif structures were within 0.67 Å RMSD of each other. Each edge represents a similarity of at least four residues. (B) The structure similarity network is shown for the three classical Rossmannoid fold subsets; a fold is a column of PDB representatives in boxes: c.2, NAD(P)-binding Rossmann-fold domains; c.3, the FAD/NAD(P)-binding domain; and c.4, the nucleotide-binding domain. These classical Rossmannoids are the most densely connected fold clique of the similarity network in (A). (C) The structure similarity network is shown for the cases when at least seven residues are shared between the two proteins. The classical Rossmannoids are well-connected, and the remaining connections are to two different Rossmannoid folds (c.23 and c.26), as well as the connection between the anti-codon-binding domain of a Class II aminoacyl-tRNA synthetase (c.51) and c.23, the flavodoxin-like fold (FixJ structure). c.26 is the adenine nucleotide α-hydrolase-like fold, which includes the Class I aminoacyl-tRNA synthetase catalytic core; the representative in the c.26 box (1iq0) is the Class I argininyl-tRNA synthetase. All structures in 1C except the two highlighted by bold boxes possess at least partial representation of the core identified in the Rossmannoids and described further in this work.