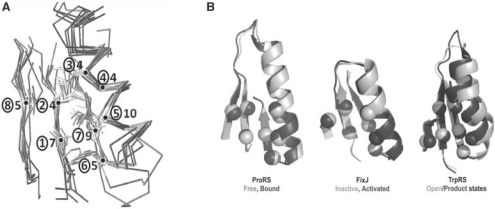
Fig. 2.
Structurally similar cores in the ProRS anti-codon-binding domain, the FixJ receiver and other Rossmannoid representatives. (A) Cores from Figure 1C and Table 1 are shown as superimposed Cα traces and stick figures for residue side chains. The sequential β−α−β motif is formed from the central and rightmost strands and the α-helix. The number of consensus amino acids is given for each residue position (circled). (B) Functionally relevant allosteric changes in ProRS, FixJ and TrpRS core motifs. These three motifs exhibit conformational changes in response to ligand binding [ProRS anti-codon-binding domain, 1H4S, 1H4T; TrpRS catalytic domain, 1MAW(F), 1I6L] or phosphorylation [FixJ, 1D5W(A), 1DBW(A)]. Activated conformations are darker. The eight positions indicated in Table 1 are indicated by Cα positions that rearrange similarly in the three proteins. These changes have been implicated experimentally in long-range communication with the active site catalytic Mg2+ ion in TrpRS (Kapustina et al., 2007; Weinreb et al., 2009).