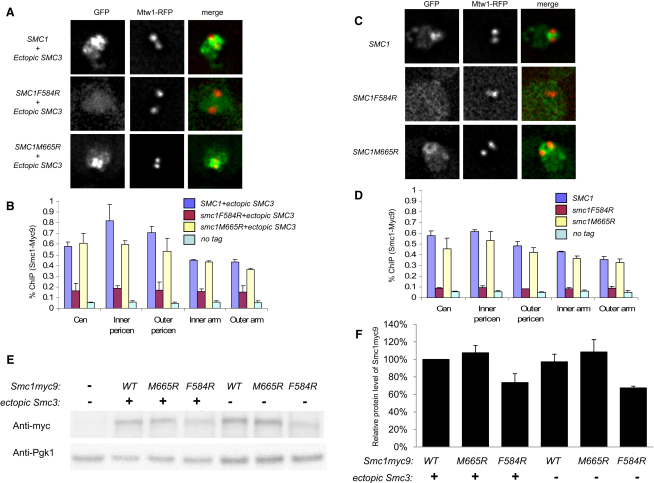
Figure 4.
Chromatin Association of Hinge Mutants
(A) Live-cell imaging shows the localization of GFP-tagged Smc1 in yeast strains with ectopically expressed Smc3. Kinetochores are marked by RFP-tagged Mtw1 to detect the spindle axes. Wild-type Smc1-GFP (K16942: MATa, SMC1-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) and Smc1(M665R)-GFP (K16944: MATa, smc1(M665R)-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) are predominantly localized in the pericentromeric region of chromosomes and appear as barrel-shaped structures. Smc1(F584R)-GFP (K16891: MATa, smc1(F584R)-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) is enriched in the nuclear region but does not form any structure.
(B) ChIP-qPCR was performed for the quantitative measurement of chromatin association of 9× Myc-tagged Smc1 at known cohesin binding loci in the presence of an ectopic copy of Smc3. Crude extracts were prepared from asynchronous cultures K14133 (MATa, SMC1-Myc9::URA3, SMC3-HA3::LEU2), K14134 (MATa, smc1(F584R)-Myc9::URA3, SMC3-HA3::LEU2), K14137 (MATa, smc1(M665R)-Myc9::URA3, SMC3-HA3::LEU2), and K699 (no tag). Error bars represent standard deviation (SD); n = 2.
(C) Live-cell imaging for Smc1-GFP localization as in (A) without an ectopic copy of Smc3. Strains used were K16428: MATa, SMC1-GFP::URA3, MTW1-RFP::KanMX; K16642: MATa, smc1(M665R)-GFP::URA3, MTW1-RFP::KanMX; and K16611: MATa, smc1(F584R)-GFP::URA3, MTW1-RFP::KanMX. Wild-type Smc1 and Smc1 M665R still form the pericentromeric barrel structure, whereas Smc1 F584R does not enrich in the nucleus.
(D) Similar experiment as in (B), but with strains without an ectopic copy of Smc3. Crude extracts were prepared from the strains K11850 (MATa, SMC1-Myc9::URA3), K13858 (MATa, smc1(F584R)-Myc9::URA3), K13860 (MATa, smc1(M665R)-Myc9::URA3), and K699 (no tag). Error bars represent SD; n = 2. (See also Figure S4.)
(E) Yeast strains K699 (no tag), K14133 (MATa, SMC1-Myc9::URA3, SMC3-HA3::LEU2), K14134 (MATa, smc1(F584R)-Myc9::URA3, SMC3-HA3::LEU2), K14137 (MATa, smc1(M665R)-Myc9::URA3, SMC3-HA3::LEU2), K11850 (MATa, SMC1-Myc9::URA3), K13858 (MATa, smc1(F584R)-Myc9::URA3), and K13860 (MATa, smc1(M665R)-Myc9::URA3) were grown to exponential phase and lysed. To determine the protein levels of Smc1-Myc9 and endogenous Pgk1, 1 or 0.1 μg of total protein was loaded onto a SDS-PAGE gel. Smc1-Myc9 and endogenous Pgk1 were detected with anti-Myc antibody and anti-Pgk1 antibody. The chemiluminescence signal was detected by the ChemiDoc XRS+ system.
(F) The density of bands in (E) was quantitated by Quantity One 1-D Analysis Software. The protein level of Smc1-Myc9 was calculated as a ratio of the density of the Smc1-Myc9 band to the density of the corresponding Pgk1 band. The protein level of Smc1-Myc9 in strain K14133 was set as 100%. Error bars represent SD; n = 3.