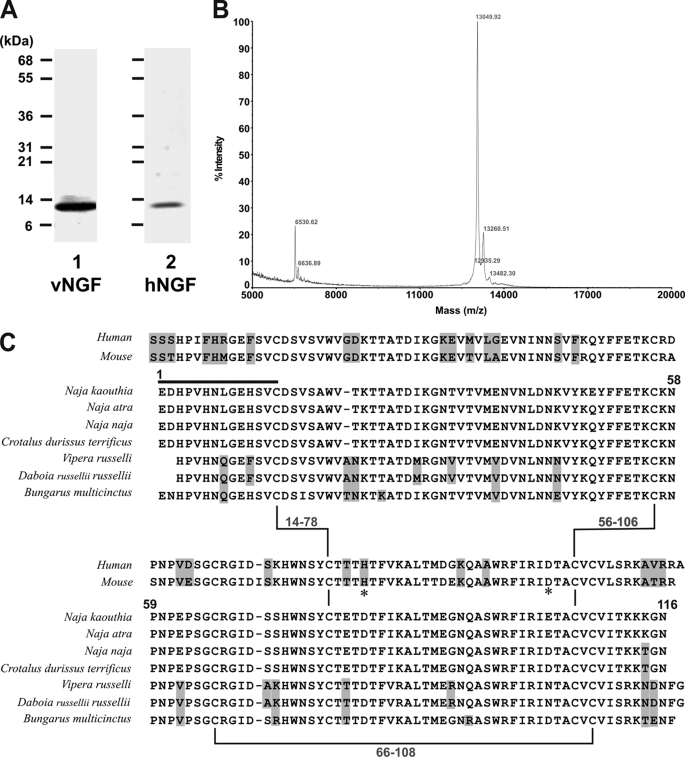
FIGURE 2.
Characterization of snake venom NGF. A, SDS 5–20% polyacrylamide gel (reducing conditions) stained with Coomassie Blue of venom NGF (vNGF, lane 1) from N. kaouthia venom or recombinant human NGF (hNGF, lane 2). B, analysis by matrix-assisted laser desorption/ionization-time of flight mass spectrometry reveals an abundant species with mass of 13,049. Peaks at 13,260 and 13,482 presumably represent single (“+206”) or double (“+412”) sinapinic acid matrix adducts within acceptable limits of accuracy. A minor peak at 6,530 (with no corresponding adduct at +206) may indicate the presence of a minor low abundance protein, and the peak at 6,636 may indicate a doubly charged species, although further analysis was not performed in detail. No higher mass peaks were detected up to 40,000 (not shown). C, comparison of human NGF, mouse NGF, and venom NGF from N. kaouthia or other species. Residue numbers refer to the N. kaouthia NGF, the black line is the N-terminal sequence determined in this study, and highlighted residues are non-identical or non-conserved in N. kaouthia NGF. Zn2+-binding sites in murine NGF involving residues His84 and Asp105 (33) are marked by an asterisk. Accession numbers/gi numbers are: human NGF, 1SG1B/49258981; mouse NGF, P01139; N. kaouthia, P61899/48429019; Naja atra, A58566/7438538; Naja naja, P01140/128164; Crotalus durissus terrificus, AAG30924/11120560; Vipera russelli, AAA03282/263157; Daboia russellii russellii, P30894/400499; Bungarus multicinctus, AAB25729/266299.