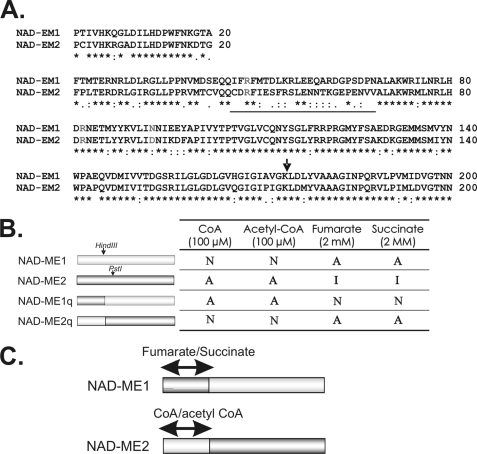
FIGURE 4.
Chimeric NAD-MEs constructed and analyzed in the present work. A, sequence alignment of the amino-terminal end of NAD-ME1 and -2 is shown. The sequences correspond to the first 200 amino acids of the mature NAD-ME1 and -2 obtained after eliminating the predicted mitochondrial targeting peptide (ARAMEMNON). The region with the most significant differences is underlined. The amino acid residues homologous to the residues involved in fumarate activation of human NAD(P)-ME and A. suum NAD-ME are indicated in light gray (33, 41, 42). The arrow indicates the site where the sequences of NAD- ME1 and -2 were exchanged for the generation of the chimeric proteins NAD-ME1q and -2q. B, shown are the regulatory properties of NAD-ME1q and -2q. The restriction sites HindIII and PstI of the parental enzymes (NAD-ME1 and NAD-ME2) were used to construct the reciprocal chimeric enzymes. The modulation of activity by CoA, acetyl-CoA, fumarate, and succinate of the parental and chimeric NAD-MEs are indicated on the right. A, activated; I, inhibited; N, no effect. C, shown are the postulated regions in the NAD-ME1 and -2 primary structure involved in allosteric activity modulation. Postulated CoA/acetyl CoA and fumarate/succinate binding sites in the parental NAD-ME sequences are indicated.