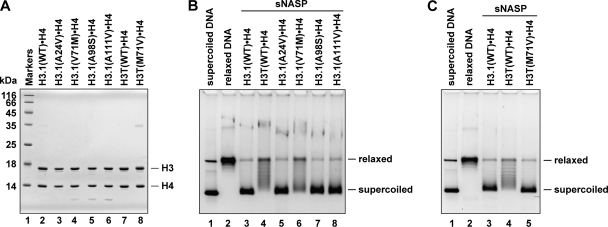
FIGURE 6.
Mutational analyses of the amino acid residue(s) required for H3T incorporation into nucleosomes. A, shown is SDS-PAGE analysis of the H3.1 and H3T mutants complexed with H4. Lane 1, molecular mass markers. Lanes 2–6, H3.1 mutants complexed with H4; lane 2, H3.1(WT); lane 3, H3.1(A24V); lane 4; H3.1(V71M); lane 5; H3.1(A98S); lane 6; H3.1(A111V). Lanes 7 and 8, H3T mutants complexed with H4; lane 7, H3T(WT), lane 8, H3T(M71V). B, nucleosome reconstitution with the H3.1 mutants by sNASP is shown. The topological assay was employed. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with sNASP in the presence of core histones. Lanes 3–8 indicate experiments with H3.1(WT)·H4, H3T(WT)·H4, H3.1(A24V)·H4, H3.1(V71M)·H4, H3.1(A98S)·H4, and H3.1(A111V)·H4, respectively. The reaction products were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer. The sNASP concentration was 1 μm. C, nucleosome reconstitution with the H3T(M71V) mutant by sNASP is shown. The topological assay was employed. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with sNASP in the presence of core histones. Lanes 3–5 indicate experiments with H3.1(WT)·H4, H3T(WT)·H4, and H3T(M71V)·H4, respectively.