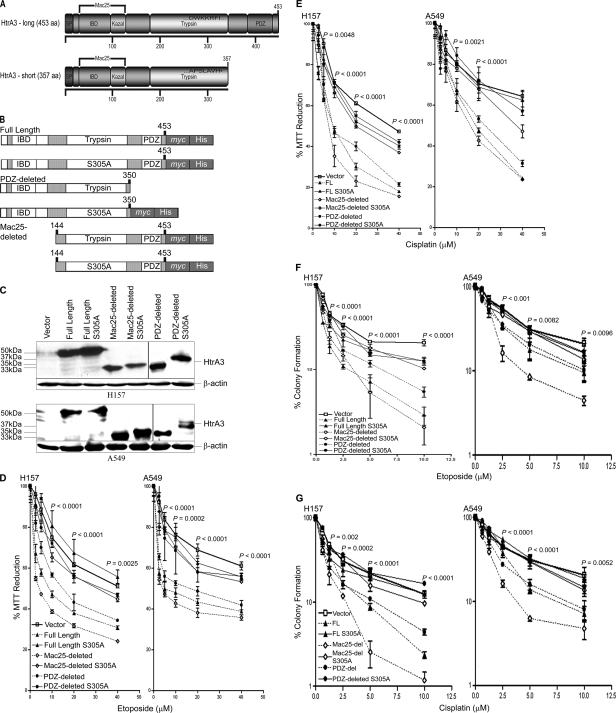
FIGURE 1.
WT HtrA3 variants modulate cytotoxicity to etoposide and cisplatin with exogenous expression in lung cancer cell lines. A, domain structural schematics of the long and short HtrA3 splice variants. The long variant is 453 amino acids in length and has an N-terminal signal peptide (SP, amino acids 1–17), an insulin/insulin-like growth factor-binding domain (amino acids 29–94), a Kazal-type serine protease-inhibitor domain (amino acids 89–126), a trypsin protease domain (amino acids 176–341), and one C-terminal PDZ domain (amino acids 384–440). The insulin/insulin-like growth factor-binding domain (IBD) and Kazal domains are homologous to Mac25. The short isoform is 357 amino acids in length and is identical to the long isoform with two exceptions: the short isoform lacks the PDZ domain, and the last seven C-terminal residues in the short form (APSLAVH) differ from the corresponding residues in the long isoform (DWKKRFI). B, domain structural schematics of WT and protease inactive full length, PDZ-deleted, and Mac25-deleted HtrA3 variants that are C-terminally myc- and polyhistidine-tagged. The WT PDZ-deleted variant was not tagged. Protease inactive HtrA3 variants contain a serine to alanine point mutation at residue 305 (S305A) within the conserved trypsin catalytic triad. C, immunoblots showing expression of HtrA3 variants, vector control, and β-actin loading controls in HtrA3-deficient lung cancer cell lines H157 and A549. PDZ-deleted variants were detected using an HtrA3 polyclonal antibody. D and E, MTT survival assays showing statistically significant attenuation of MTT reduction (p < 0.05) with exogenous expression of WT HtrA3 variants during etoposide (D) or cisplatin (E) treatment. The data were expressed as the means ± S.E. and represented at least two independent trials performed in quadruplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA. F and G, clonogenic survival assays showing decreased clonogenic survival with exogenous expression of WT HtrA3 variants following etoposide (F) or cisplatin (G) treatment. The data are expressed as the means ± S.E. and represent independent trials performed in triplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA.