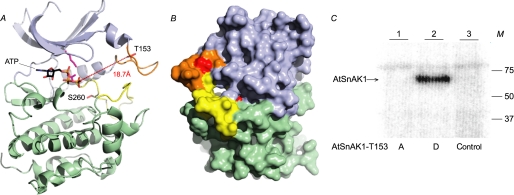
FIGURE 7.
Structural model of AtSnAK1 and accessibility of Ser260. This model was built by using the ROCK1 structure (PDB code: 2ESM, chain B) as template. A, AtSnAK1 protein as a schematic representation. The small lobe is in blue and large lobe in green, autophosphorylation (AP) loop is in orange and feedback phosphorylation (FP) loop in yellow. ATP is positioned according to the structural alignment with MEK1 (PDB code: 3E8N) and shown as lines. The side chains of interesting residues are represented in stick form (Thr153 and Ser260 in red). Distance between Thr153, and the γ-phosphate of ATP is visualized in hatched red and indicated in Å. B, protein surface model (opposite face compared with A). C, mutated AtSnAK1 proteins (2 μg of preparation) (T153A or T153D) incubated in the presence of 2 μg of AtSnRK1.1 T175D (constitutively active form) preparation in the reconstituted assay containing 2 μCi of [γ-32P]ATP and 20 μm ATP, at 30 °C for 60 min. Proteins were submitted to SDS-PAGE and autoradiography. M, molecular mass in kDa. The results are representative of three independent experiments.