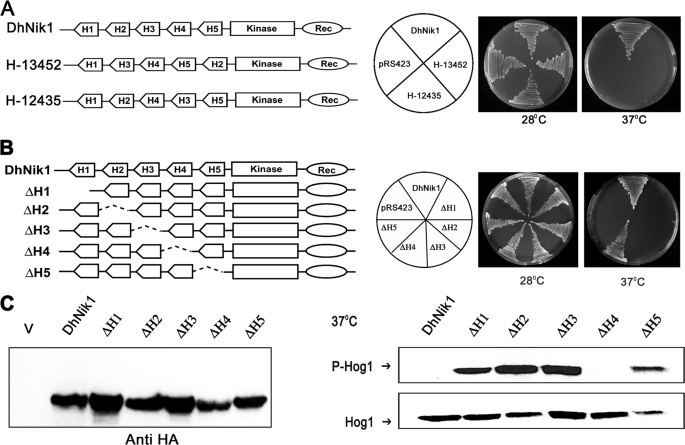
FIGURE 3.
Functional analysis of reshuffled and individually deleted HAMP domain mutants of DhNik1p. A, diagrammatic representations of DhNik1, HAMP domain reshuffled mutants H13452, or H12435 are shown in the left panel. HAMP domains are named H1, H2, H3, H4, and H5 according to their positions in DhNik1. To determine the growth patterns of TM229 harboring vector (pRS423), DhNik1, H13452, or H12435, cells were streaked on minimal SD agar plate without histidine and incubated at 28 or 37 °C for 3 days. Photographs of the respective plates are shown. B, diagrammatic representations of single HAMP domain deletion mutants and their growth patterns on minimal SD agar plate without histidine at 28 or 37 °C are shown. Experiments were repeated three times with similar results. C, left panel, immunoblot showing the level of expression of DhNik1p and HAMP-deleted mutant ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5. Total cell extracts from TM229 expressing HA-tagged DhNik1 and different mutants were immunoblotted with anti-HA antibody (Cell Signaling). Total cell extract from TM229 transformed with pRS423 were loaded as control (V). Right panel, immunoblots showing the phosphorylated Hog1p in cells with different HAMP domain deletion mutants. S. cerevisiae strain TM229 (sln1-ts) carrying DhNik1 or different mutants (ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5) were grown on SD minimal media without histidine at 28 °C until an A600 ∼0.7 and exposed to 37 °C for 1 h. Total protein extract from these cells was analyzed by Western blotting. P-Hog1 or Hog1 indicate phosphorylated or total Hog1p in the blot. Representative blots of three different experiments are shown.