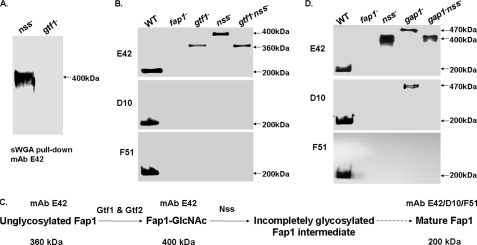
FIGURE 2.
Nss-mediated Fap1 glycosylation occurs after Gtf1-Gtf2-catalyzed glycosylation and prior to Gap1-mediated Fap1 biogenesis. Fap1 glycosylation and production profile by the nss mutant and two double mutants, gtf1-nss and gap1-nss. A, Fap1 glycosylation profiled by sWGA pull-down assays. 400 μg of whole cell lysates proteins prepared from the nss mutant (nss−) and a gtf1 mutant, VT508 (gtf1−) were subjected to sWGA pull-down and Western blotting analyses using Fap1 peptide-specific antibody mAb E42. B, Fap1 production profiled by Western blotting analysis of the gtf1 nss double mutant and its parent strains. The whole cell lysates prepared from the same number of bacterial cells (1 × 108) of wild type (WT), fap1 mutant (fap1−), VT508 (gtf1−), nss mutant (nss−), and VT508::Δnss (gtf1−nss−) were probed with Fap1-specific mAbs. C, a schematic representation of the proposed Fap1 processing intermediates. The 360-kDa unglycosylated Fap1 protein and the 400-kDa GlcNAc-modified Fap1 intermediate are recognized by peptide-specific Fap1 antibody mAb E42. The mature 200-kDa Fap1 protein reacts with all peptide- and glycan-specific Fap1 antibodies mAb E42, D10, and F51. D, Fap1 production profile by the gap1 nss double mutant and its parent strains. The whole cell lysates prepared from the same number of bacterial cells (1 × 108) of wild type (WT), fap1 mutant (fap1−), nss mutant (nss−), VT324 (gap1−), and VT324::Δnss (gap1−nss−) were subjected to Western blotting analysis using Fap1-specific mAbs.