FIGURE 1.
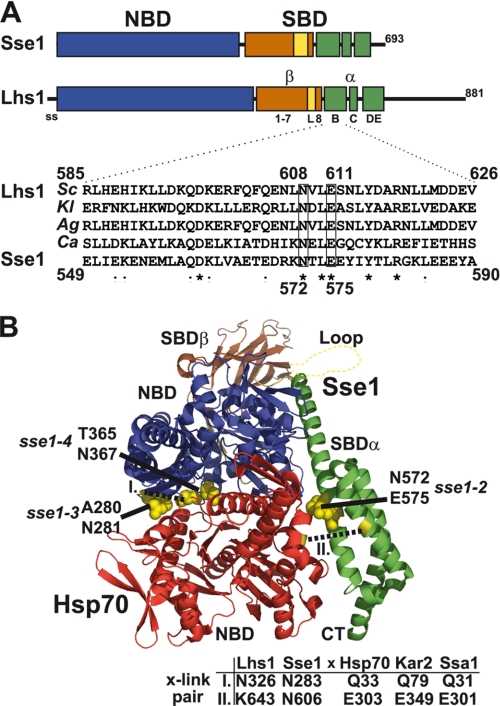
Lhs1 shares subdomain organization with Sse1. A, model for Lhs1 domain (NBD, SBD) and subdomain organization compiled from sequence alignment, local homology searches, and secondary structure prediction of Sse1 and fungal Lhs1 homologues. Indicated are the signal sequence (ss, amino acid residues 1–20), NBD (21–433, blue box), SBDβ sheet 1–7 (441–547, brown box), SBDβ loop (L, 548–564, yellow box), SBDβ sheet 8 (565–577, brown box), SBDα helix B (582–627, green box), SBDα helix C (635–650, green box), and SBDα helix DE (663–706, green box). Lhs1 has an extended C terminus with no significant homology to Sse1-(708–881). Homologous domains and subdomains of Sse1 are according to Ref. 13. Inset displays a sequence alignment of SBDα helix B from four fungal Lhs1 (S. cerevisiae, SC; K. lactis, KL; A. gossypii, Ag; C. albicans, Ca) and Sse1. Lhs1 Asn608 and Glu611 (boxed) exhibit sequence conservation with corresponding Sse1 residues (Asn572 and Glu575) that contact the Hsp70 NBD and are required for NEF activity (8). B, structure of a complex between Sse1 and the Hsp70 NBD (8). Sse1 is colored as in A and the Hsp70 NBD is colored red. Sse1 residues Thr280/Asn281, Thr365/Asn367, and Asn572/Glu575 that contact the Hsp70 NBD are marked (yellow). Residues used for pairwise cross-linking in Fig. 4 are marked (yellow, dotted lines) and corresponding positions in Lhs1/Sse1 and Hsp70/Kar2/Ssa1 are as indicated (table).