Fig. 2.
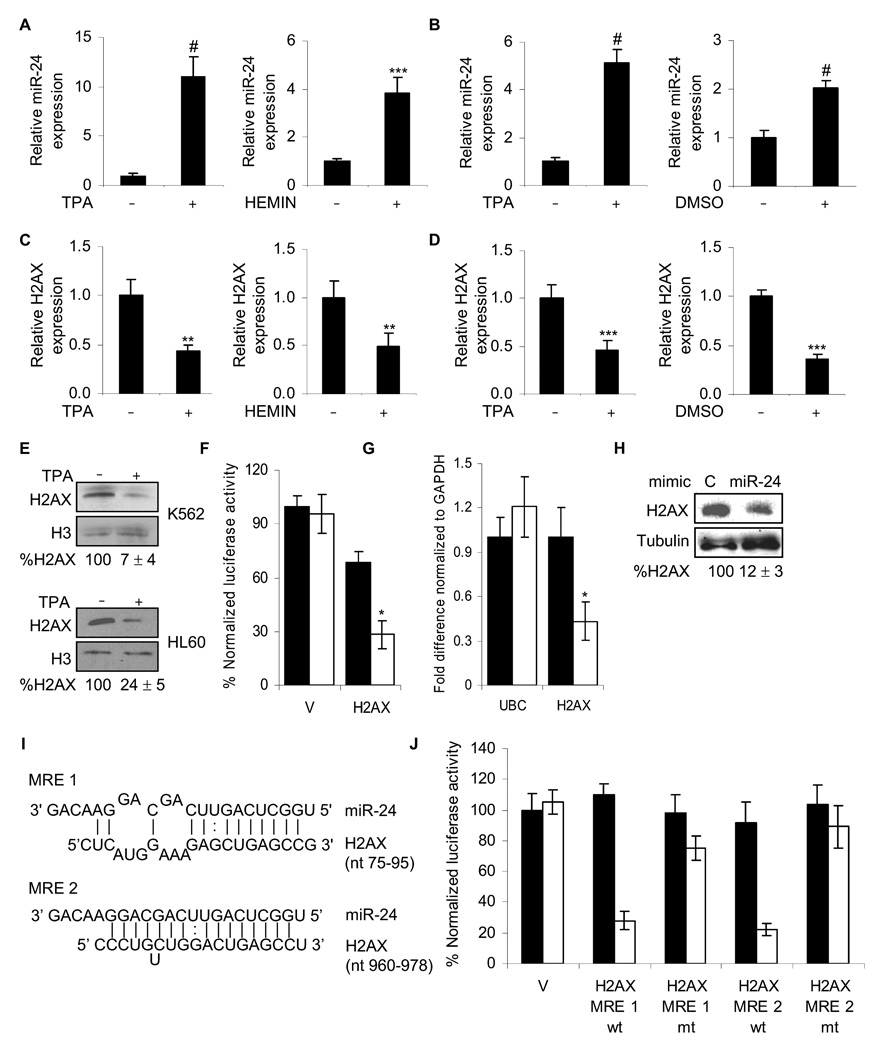
miR-24 down-regulates H2AX expression during terminal differentiation. miR-24, analyzed by qRT-PCR relative to U6, increases during differentiation of K562 cells (A) with TPA to megakaryocytes or hemin to erythrocytes (#, p< 0.001, ***, p<0.005) and during differentiation of HL60 cells (B) with TPA to macrophages or DMSO to granulocytes (#, p< 0.001). HL60 cells were treated with TPA for 2 days (left panel) or DMSO for 5 days (right panel) and miR-24 levels analyzed as described above. Under the same differentiating conditions for K562 (C) and HL60 (D) cells, H2AX mRNA, normalized to GAPDH mRNA, is down-regulated (**, p< 0.01, K562; ***, p< 0.005, HL60). (E) H2AX protein decreases after 2 d of TPA differentiation. Relative H2AX expression was quantified by densitometry using H3 as control. Lanes marked by “-” represent cells treated with vehicle alone. (F) miR-24 targets the 3’UTR of H2AX mRNA in a luciferase reporter assay. HepG2 cells were transfected with control miRNA (black) or synthetic miR-24 (white) for 48 hr and then with H2AX 3’UTR-luciferase reporter (H2AX) or vector (V) for 24 hr. Mean ± SD, normalized to vector control, of 3 independent experiments are shown (*, p< 0.001). miR-24 over-expression in HepG2 cells decreases H2AX mRNA, analyzed by qRT-PCR normalized to GAPDH (G; white, miR-24; black, cel-miR-67) and protein (H) 48 hr later. miR-24 over-expression does not alter UBC mRNA levels. (I) Schematic showing the predicted miR-24 binding sites (MRE) in the 3’UTR of H2AX mRNA. (J) Suppression of luciferase activity of a reporter gene containing in its 3’UTR the two predicted miR-24 MRE, either wild-type (wt) or with mutated seed regions (mt). HepG2 cells were transfected with control miRNA (black) or miR-24 mimic (white) for 48 hr and then with the indicated H2AX 3’UTR-luciferase reporters or vector (V). Luciferase activity was assayed 24 hr later. Mean ± SD, normalized to vector control, of 3 independent experiments are shown. In all panels, mean ± SD are shown.