Abstract
Helicases are motor proteins that are involved in DNA and RNA metabolism, replication, recombination, transcription and repair. The motors are powered by ATP binding and hydrolysis. Hepatitis C virus encodes a helicase called non-structural protein (NS3). NS3 possesses protease and helicase activities on its N-terminal and C-terminal domains respectively. The helicase domain of NS3 protein is referred as NS3h. In vitro, NS3h catalyzes RNA and DNA unwinding in a 3’ to -5’ direction. The directionality for unwinding is thought to arise in part from the enzyme's ability to translocate along DNA, but translocation has not been shown explicitly. We examined the DNA translocase activity of NS3h by using single-stranded oligonucleotide substrates containing a fluorescent probe on the 5’ end. NS3h can bind to the ssDNA and in the presence of ATP, move towards the 5’-end. When the enzyme encounters the fluorescent probe, a fluorescence change is observed that allows translocation to be characterized. Under conditions that favor binding of one NS3h per DNA substrate (100 nM NS3h, 200 nM oligonucleotide) we find that NS3h translocates on ssDNA at a rate of 46 ± 5 nt s−1 and that it can move for 230 ± 60 nt before dissociating from the DNA. The translocase activity of some helicases is responsible for displacing proteins that are bound to DNA. We studied protein displacement by using a ssDNA oligonucleotide covalently linked to biotin on the 5’-end. Upon addition of streptavidin, a ‘protein-block’ was placed in the pathway of the helicase. Interestingly, NS3h was unable to displace streptavidin from the end of the oligonucleotide, despite its ability to translocate along the DNA. The DNA unwinding activity of NS3h was examined using a 22 bp duplex DNA substrate under conditions that were identical to those used to study translocation. NS3h exhibited little or no DNA unwinding under single cycle conditions, supporting the conclusion that NS3h is a relatively poor helicase in its monomeric form, as has been reported. In summary, NS3h translocates on ssDNA as a monomer, but the translocase activity does not correspond to comparable DNA unwinding activity or protein-displacement activity under identical conditions.
INTRODUCTION
Helicases are nucleic acid –dependent ATPases that are involved in several processes such as DNA replication, repair, transcription, translation, RNA maturation, splicing and nuclear export processes (1-4). Helicases have been divided into different superfamilies such as SF1, SF2, SF3, and SF4 based on sequence analysis (5). The functional forms of helicases include oligomers (such as hexamers, trimers, or dimers) or monomers (6-10). In vitro, DNA helicases translocate along single-stranded DNA in a reaction that is coupled to ATP binding and hydrolysis. Upon encountering duplex DNA, DNA unwinding can occur, however the specific relationship between translocation and DNA unwinding has not been made clear for most helicases. Helicases that unwind DNA can displace proteins from nucleic acids while translocating along the DNA (11-14). Mutations in helicase genes lead to genomic instability which results in diseases such as Blooms syndrome, Werner syndrome, and cancer (15).
A fundamental question regarding helicase activity is the degree to which DNA unwinding is coupled to translocation activity (16). For example, DNA unwinding might be a consequence of translocation. In such a mechanism, DNA unwinding occurs due to strong interaction with one of the strands of the duplex (the translocase or bound strand) and steric displacement of the complementary strand (displaced strand) (17-19). Alternatively, specific protein-nucleic acid interactions might result in DNA melting so that DNA unwinding might be uncoupled from translocase activity (20). Dillingham et al. developed a fluorescence stopped-flow assay to study translocation on ssDNA by the B. stearothermophilus PcrA helicase (21). Later this method was modified and used by Fischer et al. to characterize the kinetic mechanism of UvrD translocation along ssDNA using global nonlinear least squares (NLLS) analysis of the kinetic time courses (22-26). These studies demonstrated that E. coli UvrD and Rep translocate progressively in 3’ to -5’ direction on ssDNA (23, 26). However, there is not a direct relationship between translocase activity and DNA unwinding for UvrD and Rep helicases. Under single cycle conditions with respect to the DNA substrate, an oligomeric form of UvrD helicase unwinds dsDNA whereas a monomer is capable of translocating along ssDNA (22). The monomeric form of UvrD does not produce ssDNA under single cycle conditions (22). Similar results have been reported for PcrA (25), and Rep (26) helicases based on global NLLS analysis, indicating that oligomerization strongly stimulates DNA unwinding activity, but is not required for translocase activity. However, a truncated form of Rep does unwind dsDNA as a monomer, and interactions between PcrA and Rep with other DNA binding proteins strongly stimulate the DNA unwinding activity in vitro (26). So the relationship of DNA unwinding to translocase activity can vary depending on the mechanism of the particular helicase, the form of the helicase, and the presence or absence of appropriate binding partners.
An activity associated with helicase translocation is the removal of proteins from DNA as a result of movement of the enzyme along the DNA (11-14). For example, PcrA and UvrD can displace RecA protein filaments from ssDNA (27, 28). A method for measuring protein displacement has been developed in which biotin labeled oligonucleotides are labeled with streptavidin to create protein blocks on the end of the DNA. Displacement of streptavidin can be measured as a result of directionally-biased movement of a helicase on ssDNA (12, 13).
Hepatitis C virus (HCV) non-structural protein 3 (NS3) is a SF2 helicase (29). The N-terminal 180 amino acids constitute a serine protease and the C-terminal 450 amino acids make up the helicase (30). NS3 catalyzes RNA and DNA unwinding in a 3’ to -5’ direction as judged by the need for a 3’ single-stranded overhang attached to the duplex (29, 30). Displacement of streptavidin from the 3’-end of biotin-labeled oligonucleotides by NS3 also supports 3’to-5’ directionality on ssDNA (14).
Full-length NS3 is capable of interacting with itself in vitro. Tackett et al. have reported that multiple NS3 molecules binding to the single stranded region of a partial duplex DNA substrate was required for optimal DNA unwinding activity in vitro (31). Sikora et al, demonstrated optimal DNA unwinding activity by an oligomeric species of NS3 based on DNA unwinding, chemical cross linking and biophysical approaches (32). Serebrov and Pyle reported that NS3 unwinds RNA as a dimer with an 18 bp kinetic step size (33). The helicase domain of NS3 is referred to NS3h, however it does not readily interact with itself based on biochemical and biophysical studies (32).
The mechano-chemical mechanism(s) of helicase stepping and coupling of ATP hydrolysis to DNA or RNA unwinding has received much attention (10, 16, 21, 23, 33, 34). Dumont et al. reported NS3 function as monomer in optical trap experiments with an 11 bp step size which was made up of smaller physical steps of 2-3 bp (35). Single molecule fluorescence energy transfer measurements supported a ‘spring-loaded’ mechanism for NS3, whereby 2-3 small steps of 1 nt are taken which builds up tension in the molecule leading to simultaneous unwinding of 2-3 bp (36). Levin et al., has studied NS3h binding and unwinding of DNA, and has concluded that the enzyme functions as a Brownian motor in which binding of ATP causes the enzyme to enter a loosely bound state in which the helicase is free to diffuse forward or backward by Brownian motion (37). Interaction with the ss/ds DNA junction facilitates ATP hydrolysis resulting in a forward ‘power-stroke’ for the proposed Brownian motor.
Optimal DNA unwinding by NS3 and NS3h requires that multiple molecules of enzyme be bound to the substrate (31, 32, 38). However, recent work indicates that a single-stranded binding (SSB) protein can substitute, at least to some degree, for the need for multiple helicase molecules because monomeric NS3h was found to be able to unwind DNA when coupled with an SSB (39). Hence, DNA and RNA unwinding by NS3 and NS3h have received much attention, and their respective mechanisms are of interest as model systems as well as in relation to hepatitis C viral replication. In contrast to DNA and RNA unwinding, translocation of NS3 or NS3h on ss nucleic acid has not been directly demonstrated.
In this report, the fluorescence stopped-flow assay using labeled oligonucleotides and global NLLS analysis were used to study translocation on ssDNA by the hepatitis C virus NS3 helicase domain. Experiments were performed under single cycle conditions that favor binding of monomeric NS3h to the DNA substrate in order to determine the rate constants, kinetic step-size, directionality and processivity of NS3h using global NLLS analysis. DNA unwinding and streptavidin displacement were also investigated under identical conditions in order to compare the different activities associated with NS3h.
EXPERIMENTAL PROCEDURES
Materials
ATP (disodium salt), Streptavidin, phosphoenol pyruvate (tricyclo-hexylammonium salt), pyruvate kinase/lactate dehydrogenase (in glycerol), heparin (sodium salt), poly (dT), Sodium dodecyl sulfate (SDS), potassium acetate, magnesium acetate, bromophenol blue, and xylene cyanol were obtained from Sigma. [γ 32p]ATP was obtained from Perkin-Elmer Life Sciences. DNA oligonucleotides were purchased from Integrated DNA Technologies and purified by denaturing polyacrylamide gel electrophoresis and stored in 10 mM Hepes (pH 7.5) and 1 mM EDTA. Purified oligonucleotides were quantified by UV absorbance at 260 nm in 0.2 M KOH by using extinction coefficients. Recombinant full length NS3 was derived from the HCV Con 1b replicon consensus sequence and was purified as described (31). NS3h was also derived from HCV Con 1B replicon consensus sequence and was purified as described (40).
Heparin was dialyzed against Milli-Q water using 3500 Da molecular mass cut-off dialysis tubing. The heparin stock solution concentration was determined by barbital buffer and titration with Azure A using previously published protocols (21-25)
Assay buffer contains 25 mM MOPS pH 7.0, 50 mM NaCl, 0.1 mM EDTA, 2 mM β-mercaptoethanol (BME), 0.1 mg/ml bovine serum albumin (BSA). For dissociation experiments the same assay buffer was prepared with out BSA.
DNA Unwinding
The substrate used to monitor NS3 and NS3h unwinding was prepared by annealing a 37mer with a 22mer resulting in a 37:22-mer partial duplex containing T15 single stranded over hang and 22 base pairs. The sequence of the loading strand was: 3’-TTTTTTTTTTTTTTTAGGACAGTCGGATCGCAGTCAG-5’ (also referred to as translocase or bound strand in some reports) and the displaced strand was 5’- TCCTGTCAGC CTAGCGTCAGTC-3’. The trapping strand was complimentary to the 22-mer displaced strand: 5’-GACTGACGCTAGGCTGACAGGA-3’. Oligonucleotides were purified by preparative gel electropheresis. Crude oligonucleotide was mixed with 90% formamide (1:1) and resolved on a 7 M urea and 20% polyacrylamide gel (Hoeffer Scientific instruments). Oligonucleotide was excised from the gel by visualizing under UV followed by electroelution of sliced gel using a Schleicher and Schull elutrap apparatus. Oligonucleotides were then desalted on a Waters Sep-Pak C18 column and lyophilized overnight using a Savant Speed-Vac. Lyophilized oligonucleotides were suspended in 10 mM Hepes (pH 7.5) and 1 mM EDTA, concentration was determined by measuring absorbance at 260 nm in 0.2 M KOH and using calculated extinction coefficients. To prepare a partial duplex substrate, 1:2 ratio of 37-mer to 22-mer was mixed, heated at 95° C for 10 min and allowed to cool to room temperature. The mixture of partial duplex was resolved on a native 20% acrylamide gel, electro eluted, desalted and quantified as described above.
The purified 37:22mer was radiolabeled at 5’-end with [γ 32p]ATP by T4 polynucleotide kinase. The substrate was incubated for 1hr at 37° C, followed by heat denaturation of the enzyme at 70° C for 10 min. Then unlabelled complimentary oligonucleotide to 22-mer was added to the 5’-radiolabelled substrate. Excess [γ 32p]ATP was removed by passing the substrate through two 1 ml Sephadex G-25 sping columns. The mixture (5’ radiolabelled and unlabelled complimentary oligonucleotide) was heated to 95° C and slow cooled to room temperature. To prepare higher concentrationsof substrate, unlabelled partial duplex DNA was added to radiolabelled substrate, heated to 95° C and slow cooled to room temperature.
DNA unwinding experiments were carried out using a KinTek rapid chemical quench-flow instrument maintained at 37° C with a circulating water bath. All concentrations listed are final concentrations after mixing. Unwinding assay buffer contained 25 mM MOPS (pH 7.0), 50 mM NaCl, 0.1mM EDTA, 2 mM BME and 0.1 mg/ml BSA. All unwinding experiments were carried out under single cycle conditions with respect to the DNA substrate by including heparin as a protein trap. For experiments in the presence of excess enzyme concentration over DNA substrate concentration, a “two-step” mixing procedure was applied. DNA (37:22mer, 2 nM) was incubated with NS3 or NS3h (500 nM) for 20 s prior to initiation of unwinding by addition of 5 mM ATP and 10 mM MgCl2. The DNA annealing trap (60 nM) and heparin (4mg/ml) were added along with ATP and MgCl2 to prevent protein rebinding to the DNA and to prevent reannealing of the ssDNA products. After rapid mixing of the solution, the reaction was quenched with 200 mM EDTA and 0.7% SDS. A 25 μl aliquot of quenched reaction mixture was added to 5 μl of loading buffer (30% glycerol, 0.1% bromophenol blue, and 0.1% xylene cyanol) and products were electrophoresed on a 20% acrylamide gel at 22 mA for 2.5 hr. The gel was exposed to a phosphor storage screen and the quantity of ssDNA and dsDNA was determined by using ImageQuant software. To determine the efficiency of heparin as a protein trap, DNA substrate (2 nM) and heparin (4 mg/ml) were incubated together prior to rapid addition of NS3h (500 nM). After 20 s, the unwinding reaction was initiated by addition of ATP (5 mM), MgCl2 (10 mM) and annealing trap (60 nM). For experiments conducted under conditions of excess substrate concentration relative to enzyme concentration, DNA (37:22mer, 200 nM) was incubated with NS3 or NS3h (100 nM) prior to initiation of unwinding with 5mM ATP and 10 mM MgCl2, 4mg/ml heparin and the DNA annealing trap (2 μM). To determine the efficiency of heparin as a protein trap under conditions of excess substrate concentration,DNA substrate (37:22mer, 200 nM) was incubated with 4 mg/ml heparin, 5 mM ATP, 10 mM MgCl2, and 2 μM DNA annealing trap prior to initiation of DNA unwinding by addition of NS3h (100 nM). The reaction was quenched, loading dye was added, and products were separated by electrophoresis and quantitated as described above.
Streptavidin Displacement
This assay was performed by using two 5’-biotinylated oligonucleotides where biotin label (Biotin dT) was incorporated into the DNA. The sequence of the 5’-bio-30mer (29 nucleotides plus Biotin dT) was 5’-GX(T)28-3’ and 5’-bio-60mer (59 nucleotides plus Biotin dT) was 5’ GX(T)58-3’ where X indicates placement of biotin label in the DNA sequence. Streptavidin (50 μM) was prepared in buffer containing 25 mM Hepes, pH 7.5, 20 % glycerol, and 10 mM NaCl. The solution was quick-frozen in liquid nitrogen and stored at -80 °C. Prior to use, streptavidin was diluted to 5 μM in buffer consisting of 25 mM Hepes, pH 7.5, 0.1 mg/ml BSA, 0.1 mM EDTA, and 1 mM BME. Oligonucleotide (10 nM or 200 nM) was incubated in reaction buffer [25 mM MOPS (pH 7.0), 12.5 mM Mg(OAc)2, 150 mM KOAc, 4 mM PEP, 1 mM BME, and 0.1 mg/ml BSA] with 5 mM ATP, 300 nM streptavidin, and PK/LDH (10.8 and 16.6 units/ml, respectively) at 37°C for 3 min. For experiments which utilized 200 nM oligonucleotide, 6 μM of streptavidin was included rather than 300 nM in order to maintain the same ratio of DNA to streptavidin in all the experiments. The streptavidin displacement reaction was initiated by addition of helicase. At various times, 10 μl aliquots were removed from the reaction and mixed with 10 μl of quench solution [0.6 % SDS, 200 mM EDTA, pH 8.0, 10 uM poly(dT), 0.08% xylene cyanol, 0.08% bromophenol blue, and 10% glycerol]. Samples were analyzed by electrophoresis on a 15% polyacrylamide gel. Radiolabeled DNA bands were visualized by using a phosphorimager and the quantity of radioactivity in the bands corresponding to free oligonucleotide and streptavidin-bound oligonucleotide in each sample was determined by using ImageQuant software. The fraction of free oligonucleotide in samples was calculated with a correction for the free oligonucleotide in the blank sample by using the following equation.
FDc,t is the fraction of free oligonucleotide corrected for the amount of free oligonucleotide in the blank sample. FDt is the radioactivity of free oligonucleotide DNA for each sample at time t. SDt is the radioactivity of streptavidin-bound oligonucleotide DNA at time t. FDb and SDb are the radioactivity of free oligonucleotide DNA and streptavidin bound oligonucleotide DNA, respectively in the blank (b) at time zero.
Dissociation of NS3h from ssDNA
Dissociation experiments were performed as described previously (22, 25). Briefly, dissociation kinetics were determined by an increase in NS3h tryptophan fluorescence (λex = 280 nm, and λem = 345 nm). The dissociation rate constant (kd) was measured during NS3h translocation on ssDNA (poly (dT)).
Translocation of NS3h on ssDNA
Translocation experiments were performed in assay buffer (25 mM MOPS (pH 7.0), 50 mM NaCl, 0.1mM EDTA, 1 mM β-mercaptoethanol, and 0.1 mg/ml bovine serum albumin) at 37 °C using a SX18MV stopped-flow instrument [Applied Photophysics Ltd. Surrey, UK]. During translocation experiments, 100 nM NS3h was preincubated with 200 nM ssDNA and reaction was initiated by the addition of 5 mM ATP, 10 mM MgCl2, and 4 mg/ml heparin. These are final concentrations after mixing in the stopped- flow instrument. NS3h translocation was measured as described previously (22). Briefly fluorescence change was measured upon NS3h arrival at the 5’ end of ssDNA using 5’-F-(dT)L (λex = 492 nm, λem = >520 nm).
Analysis of Kinetic Translocation Data
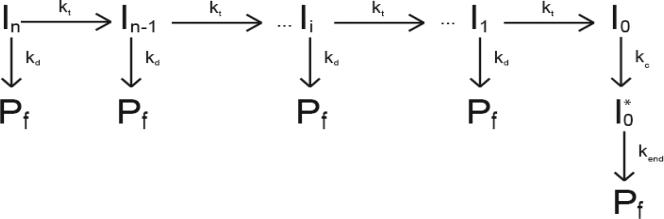
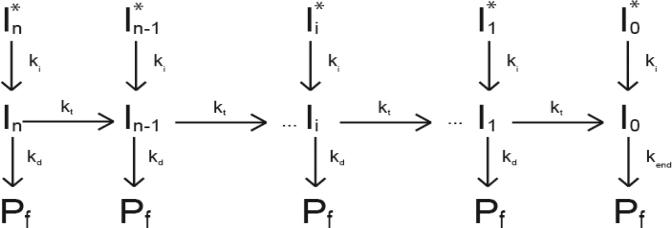
The multiple time courses for NS3h monomer translocation on DNA, each corresponding to a different length of DNA, were analyzed globally using equation (1) (derived from Scheme 1) or equation (2) (derived from Scheme 2) and standard non-linear least squares (NLLS) algorithms to obtain estimates of the translocation kinetic parameters (22-26). In these equations the variables kt, kd, kc, ki, kend, m, d, B, C, r and rn were constrained to be global parameters (independent of DNA length); the A variables were allowed to float for each time course (23-26). The multiple time course for NS3h monomer translocation on poly(dT), each corresponding to a different solution concentration of heparin, were analyzed separately using equation (3). All NLLS and linear least squares analyses were performed using Conlin (41) kindly provided by Dr. Jeremy Williams. The software library CNL50 (Visual Numerics Incorporated, Houston, TX) was used for the numerical calculation of the inverse Laplace transform.
Scheme I.
Scheme II.
RESULTS
DNA unwinding under conditions of enzyme concentration in excess of substrate concentration
In order to investigate the relationship between translocation on ssDNA and unwinding of dsDNA for NS3h and for NS3, conditions were sought that could be applied to each type of measurement and to each form of the enzyme. NS3 exhibits a large kinetic step size as well as relatively low processivity. A DNA substrate was prepared that would be short enough to exhibit a single kinetic step (and therefore more product), but was stable enough to use at 37 °C. The substrate contained 15 nt of ssDNA overhang and 22 base pairs (37nt:22bp). In order to evaluate the substrate, standard DNA unwinding experiments were performed in which the helicase concentration exceeded the DNA substrate concentration. Typical experiments for measuring DNA unwinding by a helicase are conducted by incubating the helicase with the DNA substrate, followed by rapid mixing with ATP and Mg+2 to initiate the reaction. However, we found that this protocol does not work well with NS3h when the helicase concentration is in excess of the DNA substrate concentration due to ATP-independent DNA unwinding (42).
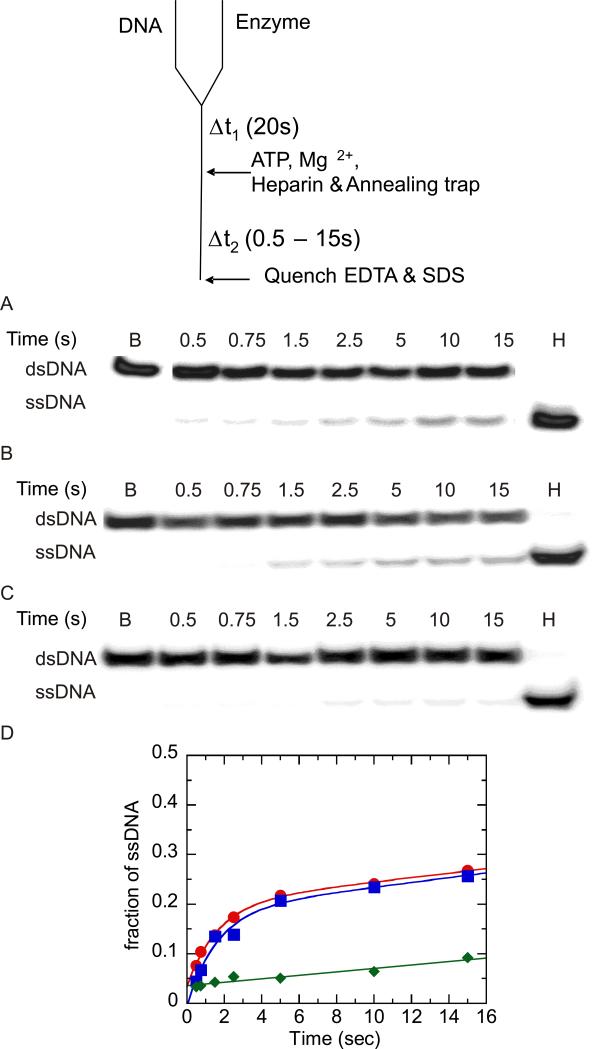
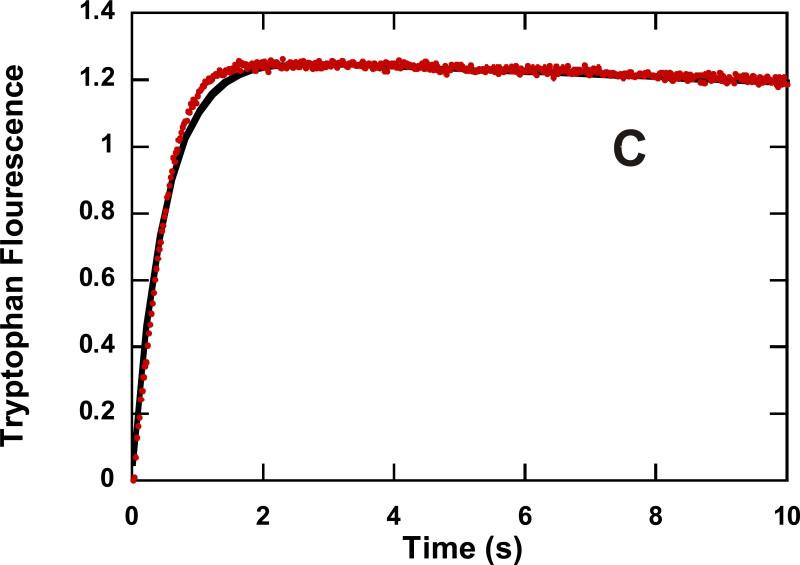
DNA unwinding by NS3h has been studied extensively by using a “double-mixing” approach as described by Levin et al. (38). We applied this approach to compare DNA unwinding by NS3h and NS3 under identical conditions (Figure 1). Helicase is rapidly mixed with DNA substrate and allowed to incubate for 20 s, which is too little time for ATP-independent melting to occur. After the 20 s incubation time, ATP and Mg+2 are introduced in a second rapid mixing step, followed by varying reaction times before stopping the reaction with quencher. Using this approach, we were able to directly compare DNA unwinding by NS3 and NS3h under identical conditions. NS3 or NS3h (500 nM) was rapidly mixed with DNA (2 nM) followed a second rapid mixing step in which ATP and MgCl2 were added. To prevent reannealing of ssDNA products, a 22-mer DNA annealing trap was included with the ATP and MgCl2, which was complimentary to the 22-mer of the 37:22-mer DNA substrate. To maintain single cycle conditions, heparin was also added to ‘trap’ protein that dissociated from the DNA substrate. The efficiency of heparin as a protein trap was tested by adding heparin to the DNA substrate prior to the first mixing step. Under these conditions a small amount of DNA unwinding was observed, which indicated heparin as a good, but not perfect protein trap under these conditions (Figure 1C). NS3 and NS3h unwind DNA almost identically, producing around 20% ssDNA product when incubated with substrate for only 20 s prior to initiation of the unwinding reaction (Figure 1D). In contrast, NS3 unwinds nearly 80% of a 30 bp DNA substrate when incubated for at least 10 min prior to initiation of the reaction (29-31, 40). Therefore, the incubation time between NS3 and DNA can affect the level of DNA unwinding, as has been reported for NS3 unwinding of RNA (43). The slow phase for DNA unwinding following the initial burst of product formation is likely due to slight leakage of the protein trap based on similar rates for the slow phase compared to the control experiment (Figure 1D).
Figure 1.
Unwinding of 37:22-mer partial duplex by NS3 and NS3h in the presence of heparin (used as a protein trap) under conditions of enzyme concentration in excess of DNA substrate concentration. A “double-mixing” experimental protocol was performed as described in the text. Unwinding products were resolved on 20% polyacrylamide gel and visualized using a PhosphorImager and quantitated by using ImageQuant software. (A) Representative gel image showing unwinding of 2 nM partial duplex by 500 nM NS3. The blank sample (B) shows the partial duplex DNA substrate prior to unwinding, and sample (H) shows the DNA after heating to 95 °C for ten min. (B) Representative gel image showing unwinding of 2 nM partial duplex by 500 nM NS3h. (C) To determine the efficiency of heparin as a protein trap, the partial duplex substrate (2 nM) was mixed with heparin (4 mg/ml) prior to initiating the reaction upon mixing with NS3h (500 nM). Little or no unwinding was observed. (D) Fraction of product formation for unwinding of 2 nM partial duplex by 500 nM NS3 ( ), 500 nM NS3h (
), 500 nM NS3h ( ), and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. Enzyme-catalyzed unwinding was fit to a single exponential followed by a steady state rate resulting in amplitudes of 0.19 ± .03 and 0.17 ± .01 nM, rate constants of 0.62 ± 0.3 and 0.61 ± 0.1 s−1, and steady state rates of 0.005 ± .001 and 0.004 ± 0.001 nM/s for NS3 and NS3h, respectively. The control experiment in which heparin was added to the DNA substrate prior to addition of NS3h was fit to a linear equation resulting in a rate of 0.004 ± 0.001 nM/s.(
), and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. Enzyme-catalyzed unwinding was fit to a single exponential followed by a steady state rate resulting in amplitudes of 0.19 ± .03 and 0.17 ± .01 nM, rate constants of 0.62 ± 0.3 and 0.61 ± 0.1 s−1, and steady state rates of 0.005 ± .001 and 0.004 ± 0.001 nM/s for NS3 and NS3h, respectively. The control experiment in which heparin was added to the DNA substrate prior to addition of NS3h was fit to a linear equation resulting in a rate of 0.004 ± 0.001 nM/s.( ).
).
DNA unwinding under conditions of DNA substrate concentration in excess of enzyme concentration
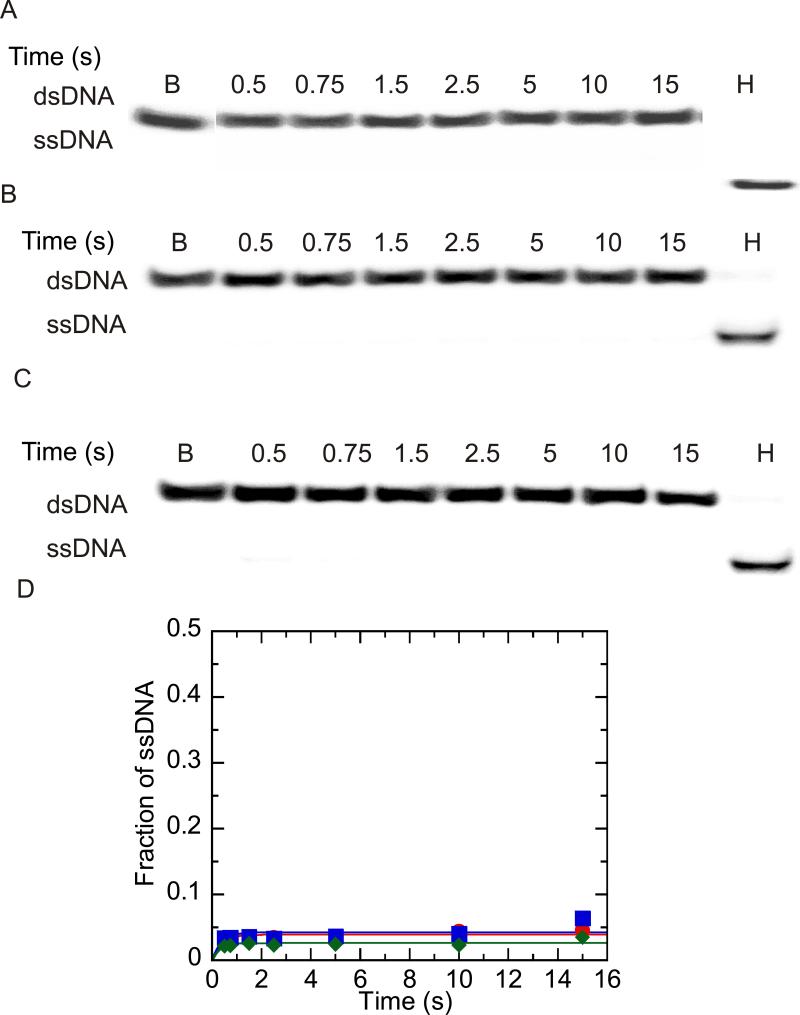
Translocation on ssDNA has been best studied under conditions that favor binding of one molecule of helicase per one molecule of substrate (16). In order to obtain these conditions, the concentration of oligonucleotide substrate must be in excess of the helicase concentration. DNA unwinding experiments were conducted under these conditions, in which the DNA substrate concentration (200 nM) exceeded the enzyme concentration (100 nM). NS3h has been shown to exist as a monomer in the presence and absence of ssDNA (32, 44 and supplemental figure 1.) When NS3h is pre-incubated with the DNA substrate under these conditions, no ATP-independent unwinding was observed (Reynolds, K.A. and Raney, K.D., in preparation). Therefore, only one mixing step was necessary for unwinding experiments to be performed. NS3 or NS3h (100 nM) and DNA (200 nM) were incubated prior to initiation of unwinding by addition of ATP, MgCl2, heparin and annealing trap (Figure 2). Under single cycle conditions little or no unwinding was observed (Figures 2A, 2B and 2D). The efficiency of the heparin as a protein trap was tested under these conditions by incubating DNA with ATP, MgCl2, heparin and annealing trap prior to initiation of unwinding with NS3h which led to no observable unwinding over background, indicating that heparin functions as an excellent protein trap under these conditions (Figures 2C and 2D).
Figure 2.
Unwinding of 37:22-mer partial duplex by NS3 and NS3h under conditions of DNA substrate concentration in excess of enzyme concentration. Unwinding products were resolved by electrophoresis on a 20 % native polyacrylamide gel, visualized using a PhosphorImager and quantitated by using ImageQuant software. (A) Representative gel image showing unwinding of 200 nM partial duplex DNA by 100 nM NS3. The blank sample (B) shows the DNA substrate prior to unwinding, and the heated sample (H) show the substrate after heating to 95 °C. (B) Representative gel image showing unwinding of 200 nM partial duplex DNA by 100 nM NS3h. (C) Heparin (4 mg/ml) was mixed with the partial duplex substrate (200 nM) prior to initiating the reaction by mixing with NS3h (100 nM) to determine the ability of heparin to serve as a protein trap. (D) Fraction of ssDNA product formed after quantitation of gel images by using Imagequant software. Unwinding of 200 nM 37:22-mer DNA by 100 nM NS3 ( ), 100 nM NS3h (
), 100 nM NS3h ( ) and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. (
) and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. ( ).
).
Protein displacement by NS3 and NS3h
Helicase activities are not limited to melting duplex DNA. Helicases can also displace proteins from DNA during DNA unwinding. Displacement of streptavidin from biotin-labeled oligonucleotides has been used to measure protein displacement activity on ssDNA. A 5’-bio-29mer and a 5’-bio-59mer were examined to compare the protein displacement activities of NS3 and NS3h on ssDNA. The reaction was initiated by the addition of NS3 or NS3h to the biotin-labeled DNA substrate (10 nM) in the presence of ATP, Mg+2, and excess biotin which served to trap streptavidin. Aliquots were removed and quenched at various times, followed by native polyacrylamide gel electrophoresis to separate free oligonucleotide from streptavidin-bound oligonucleotide (Figures 3 and 4). The gel band due to streptavidin bound oligonucleotide decreased during the 60 min incubation with NS3 for the 5’-bio-29mer and 5’-bio-59mer, indicating streptavidin displacement from biotin-labeled oligonucleotide (Figures 3A, and 3B). Similarly, streptavidin displacement reaction was also performed by the addition of 1 μM NS3h to 10 nM substrate however, no streptavidin displacement was observed in the presence of NS3h (Figures 4A and 4B).
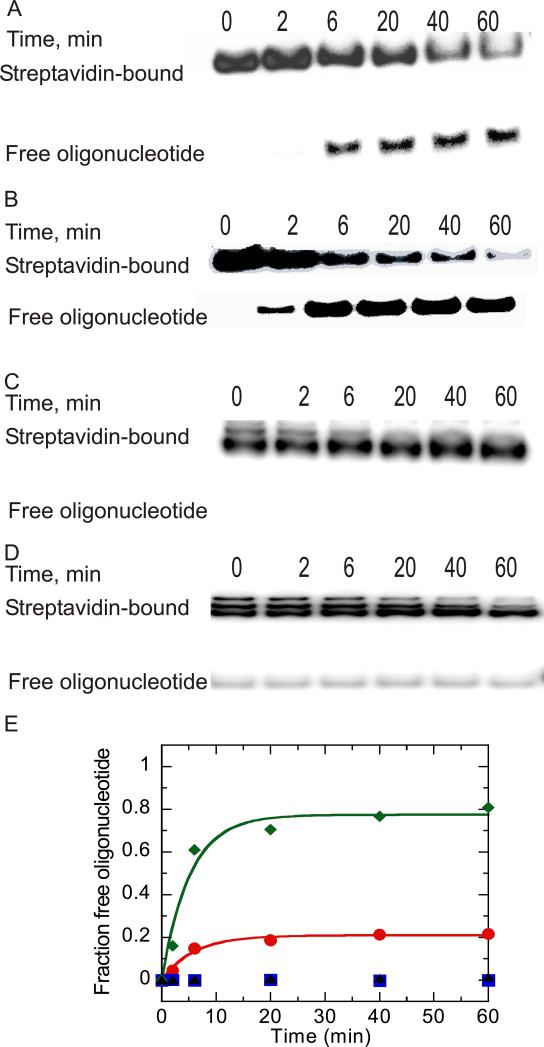
Figure 3.
NS3 catalyzed streptavidin displacement from 5’-bio-30mer and 5’-bio-60mer oligonucleotides. (A) NS3 (1 μM) was incubated with 5’-bio-30mer (10 nM) in reaction buffer. Samples were removed at varying times and added to a quencher solution (0.6% SDS, 200 mM EDTA, 0.08% Xylene cyanol, 0.08% bromophenol blue, and 10% glycerol) followed by separation of streptavidin-bound oligonucleotide from free oligonucleotide on a 15% native polyacrylamide gel. (B) NS3 (1 μM) was incubated with 5’-bio-60mer 10 nM in reaction buffer and treated as described for panel A.. (C) NS3 (100 nM) was incubated with 5’-bio-30mer 200 nM under identical reaction conditions as described for panel A, and samples were analyzed by 15% native polyacrylamide gel electrophoresis. (D) NS3 (100 nM) was incubated with 5’-bio-60mer (200 nM) under identical conditions as described for panel A. (E) Streptavidin displacement from different lengths of biotin oligonucleotides catalyzed by NS3. Results were obtained by quantitation of gels in panels A-D. Streptavidin displacement over time is shown in the presence of NS3 (1 μM) incubated with 10 nM 5’-bio-30mer ( ),NS3 incubated with 10 nM 5’-bio-60mer (
),NS3 incubated with 10 nM 5’-bio-60mer ( ), NS3 (100 nM) incubated with 200 nM 5’-bio-30mer (
), NS3 (100 nM) incubated with 200 nM 5’-bio-30mer ( ), and NS3 (100 nM) incubated with 200 nM 5’-bio-60mer (▲).
), and NS3 (100 nM) incubated with 200 nM 5’-bio-60mer (▲).
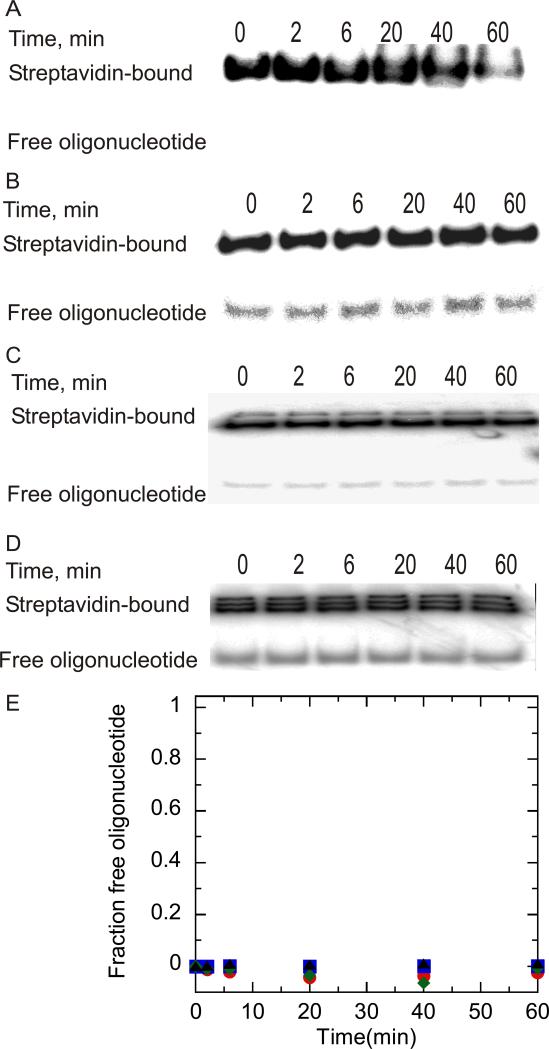
Figure 4.
NS3h catalyzed streptavidin displacement from 5’-bio-30mer and 5’-bio-60mer oligonucleotides. (A) NS3h (1 μM) was incubated with 5’-bio-30mer (10 nM) in reaction buffer (describe). Samples were removed at varying times and added to a quencher solution (0.6% SDS, 200 mM EDTA, 0.08% Xylene cyanol, 0.08% bromophenol blue, and 10% glycerol) followed by separation of streptavidin-bound oligonucleotide from free oligonucleotide on a 15% native polyacrylamide gel. (B) NS3h (1 μM) was incubated with 5’-bio-60mer 10 nM in reaction buffer and treated as described for panel A.. (C) NS3h (100 nM) was incubated with 5’-bio-30mer 200 nM under identical reaction conditions as described for panel A, and samples were analyzed by 15% native polyacrylamide gel electrophoresis. (D) NS3h (100 nM) was incubated with 5’-bio-60mer (200 nM) under identical conditions as described for panel A. (E) Streptavidin displacement from different lengths of biotin oligonucleotides catalyzed by NS3. Results were obtained by quantitation of gels in panels A-D. Streptavidin displacement over time is shown in the presence of NS3h (1 μM) incubated with 10 nM 5’-bio-30mer ( ),NS3h incubated with 10 nM 5’-bio-60mer (
),NS3h incubated with 10 nM 5’-bio-60mer ( ), NS3h (100 nM) incubated with 10 nM 5’-bio-30mer (
), NS3h (100 nM) incubated with 10 nM 5’-bio-30mer ( ), and NS3h (100 nM) incubated with 10 nM 5’-bio-60mer (▲).
), and NS3h (100 nM) incubated with 10 nM 5’-bio-60mer (▲).
In order to mimic the conditions used for translocation assays (described below) streptavidin displacement experiments were carried out by adding 100 nM NS3h to 200 nM substrate prior to the reaction. No product was observed with either substrates (5’bio-29mer or 5’-bio-59mer) during the 60 min incubation, which indicated no streptavidin displacement under these conditions (Figures 3C and 3D, 4C and 4D). In summary, NS3 was capable of displacing streptavidin from both DNA substrates under conditions in which the helicase concentration greatly exceeded the substrate concentration. However, when NS3 concentration (100 nM) was less than the substrate concentration (200 nM), no streptavidin displacement was observed during the 60 min incubation (Figure 3). NS3h was unable to displace streptavidin from either biotin-labeled oligonucleotide, regardless of the relative concentrations of enzyme and substrate (Figure 4). Hence, the DNA unwinding properties of these two helicases are similar when measured under identical conditions; however their protein displacement activities are quite different.
We have added a new experiment in which NS3h dissociation was measured on a biotin-labeled substrate in the presence or absence of streptavidin (Supplemental Figure 6). NS3h dissociates at a similar rate on each substrate. This indicates that when NS3h encounters streptavidin, it dissociates just as it would as if reaching the end of ssDNA. This result is in contrast to the result with Dda helicase, which was found to remain, bound to ssDNA upon colliding with streptavidin (45). Previous results with other helicases indicated no difference between the BioTeg and Biotin dT linkers. In this paper, we used biotin dT and we mention this in the Experimental Procedures section. However, it is theoretically possible that different linkers might cause a difference. Examination of the structural models of streptavidin bound to biotin-labeled DNA and considering the molecular structure of NS3h, it is difficult to imagine how NS3h might evade the streptavidin molecule during translocation.
NS3h is a 3’ to -5’ directionally biased single-stranded DNA translocase
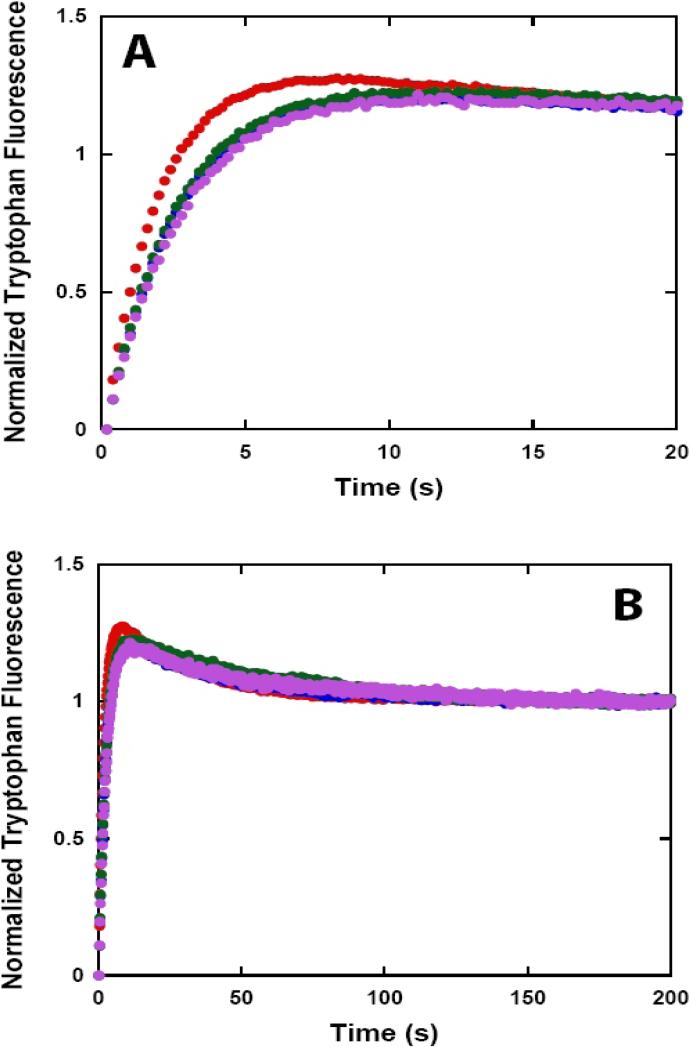
The translocation of NS3 and NS3h along single-stranded DNA was monitored in a stopped-flow spectrometer using a previously established technique (20-23). This approach measures the change in fluorescence that occurs as a helicase translocates toward or away from a fluorescent label on ssDNA. The dependence of the observed time courses of fluorescence on the length of the oligonucleotide allows one to determine the directionality and the kinetic parameters that govern translocation. We used a series of (dT)L molecules (L: 40 – 100 nt) labeled at the 3’ or 5’ end with fluorescein (5’-F-(dT)Lor 5’-(dT)L-F) since the binding of enzyme to fluorescein results in a quenching of the fluorescence of the fluorophore (Figure 6). The protein trap heparin was also included in these reactions to ensure that any free enzyme in solution at the start of the reaction and any enzyme that dissociates during translocation will be unable to rebind the DNA and reinitiate translocation (22-25).
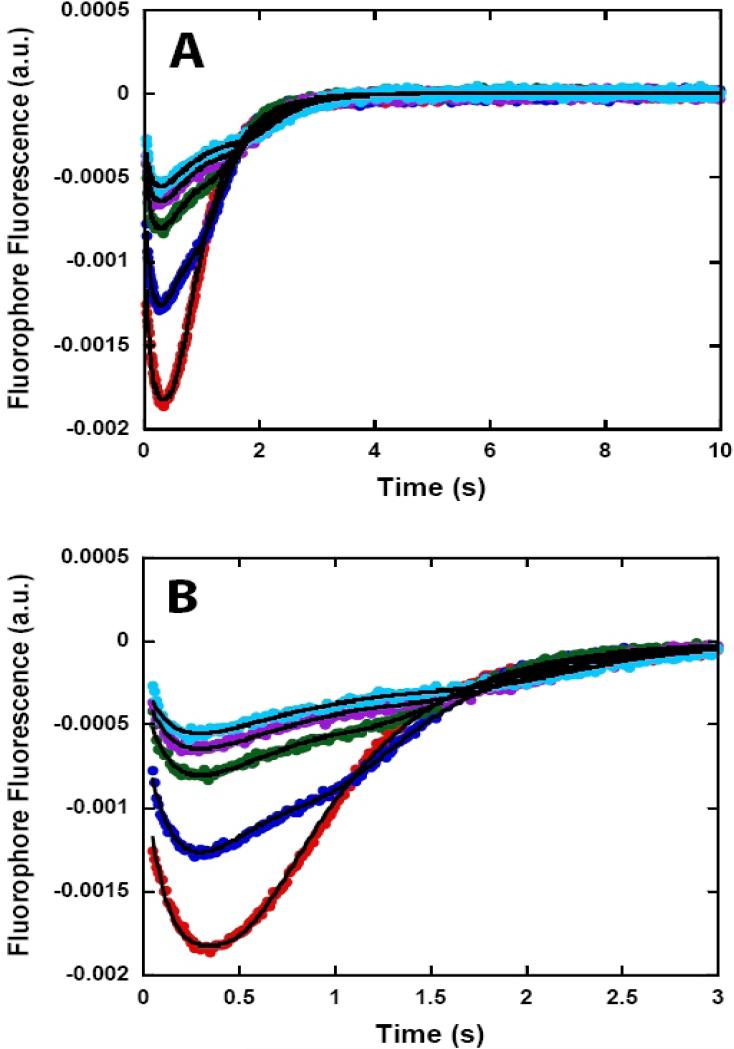
Figure 6.
NS3h translocation kinetics monitored by stopped-flow experiments with fluorescein labeled ssDNA. NS3h (100 nM) was pre-incubated with 200 nM ssDNA labeled at the 5’ end with fluorescein (5’-F-(dT)L at 37 °C in assay buffer and translocation was initiated by the addition of ATP:Mg2+ and heparin to a final concentration of 5 mM, 10 mM, and 4 mg/ml, respectively. Fluorescence time courses at short (A) and long (B) time scales obtained with 5’-F-(dT)40 ( ), 5’-F-(dT)60 (
), 5’-F-(dT)60 ( ), 5’-F-(dT)72 (
), 5’-F-(dT)72 ( ), 5’-F-(dT)88 (
), 5’-F-(dT)88 ( ) and 5’-F-(dT)100 (
) and 5’-F-(dT)100 ( ). The solid lines in the figure are simulations using equation 1 and the best fit parameters obtained from the global NLLS analysis of the time courses using equation 1.
). The solid lines in the figure are simulations using equation 1 and the best fit parameters obtained from the global NLLS analysis of the time courses using equation 1.
Multiple attempts to examine translocation of NS3 using different lengths of oligonucleotides labeled with fluorescein failed to produce interpretable data (supplemental Figure 2). NS3 can exist as an oligomer containing multiple subunits. Preliminary experiments using DNA footprinting techniques indicate that NS3 binds along the entire length of the ssDNA due to protein-protein interactions (Raney, V.M., and Raney, K.D. in preparation). This mode of binding precludes the study of translocation using the fluorescein-labeled substrates under conditions reported here, so alternative methods will be needed to examine translocation of this enzyme on ssDNA. Therefore, additional experiments were performed only with NS3h.
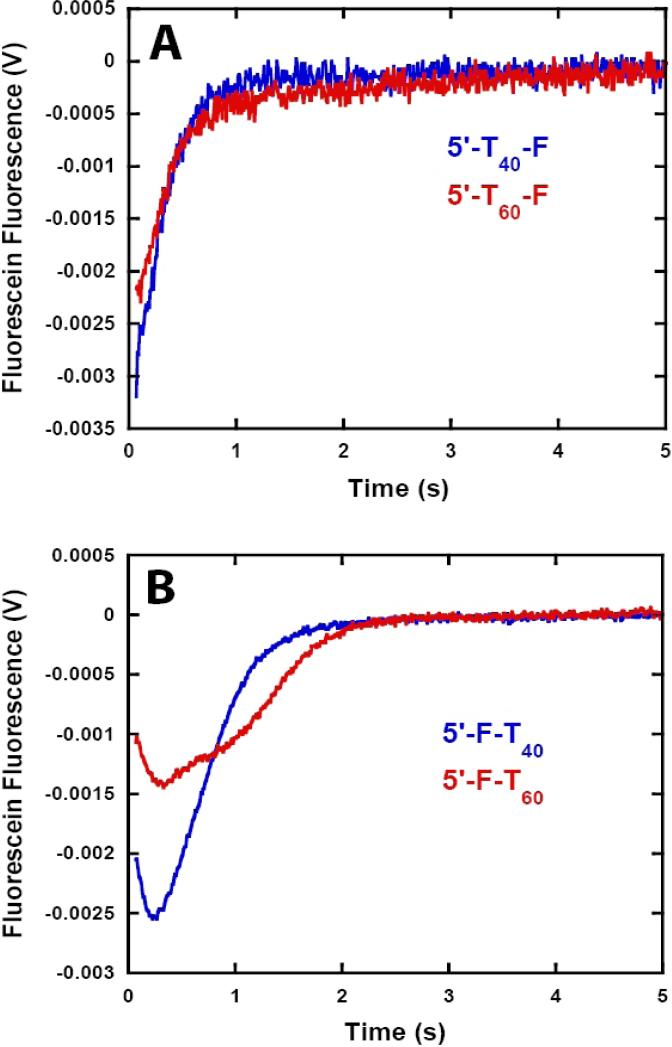
As shown in Figure 5A, when fluorescein is at the 3’ end of the ssDNA only an increase in the fluorescence of the fluorophore is observed. Furthermore, the qualitative features of the time courses were independent of the length of the DNA. In contrast, when the fluorophore is at the 5’ end of the DNA the observed time courses of fluorescence do depend upon the length of the DNA and are multiphasic; an initial rapid decrease in the fluorescence of the fluorophore is followed by a slow increase in fluorescence (Figure 5B). Taken together, these data support the conclusion that NS3h is a 3’ to -5’ directionally biased single-stranded DNA translocase (14).
Figure 5.
NS3h translocation kinetics monitored by stopped-flow experiments with fluorescein labeled ssDNA. NS3h (100 nM) was pre-incubated with 200 nM ssDNA in assay buffer and translocation was initiated by the addition of ATP (5 mM), MgCl2 (10 mM) and heparin (4 mg/ml). (A) Fluorescence time courses obtained with 5’-(dT)40–F ( ) and 5’-(dT)60-F (
) and 5’-(dT)60-F ( ). (B) Fluorescence time courses obtained with 5’-F-(dT)40 (
). (B) Fluorescence time courses obtained with 5’-F-(dT)40 ( ) and 5’-F-(dT)60 (
) and 5’-F-(dT)60 ( ).
).
Kinetic analysis of NS3h translocation along ssDNA using fluorescein-labeled DNA
The fluorescence time-courses resulting from stopped-flow experiments obtained with (5’-F-(dT)L), L = 40, 60, 72, 88, and 100 are shown in Figure 6. These experiments were carried out at 37 °C in assay buffer at a final heparin concentration of 4 mg/ml. The time courses of single-stranded DNA translocation by NS3h are qualitatively similar to those previously observed for single-stranded DNA translocation by the E. coli UvrD (22), B. stearothermophilus PcrA, (25) and E. coli Rep (26) helicases, which suggests that a similar model for single-stranded DNA translocation may be applicable. It is worth noting, however, that there is evidence of a very fast initial phase in the time courses of single-stranded DNA translocation by NS3h occurring on a timescale less than the first 50 ms of the reaction. This magnitude and duration of this fast phase, associated with a rapid increase in the fluorescence of the fluorophore, was independent of the length of the DNA, suggesting that it corresponds to a physical process carried out by proteins already bound close to the fluorophore before the addition of ATP. However, since the measurements of fluorophore fluorescence associated with these early time scales had large uncertainty, we did not attempt to model this phase directly. This omission will not affect our subsequent estimates of the kinetic parameters associated with the translocation of NS3h along single-stranded DNA since, as stated, this magnitude and duration of this phase was not dependent upon the length of the single-stranded DNA.
The model for the mechanism of single-stranded DNA translocation by NS3h is shown in Scheme 1. In this model, an NS3h monomer with an occluded site size of b nucleotides and a contact size of d nucleotides binds with polarity to a single stranded DNA, L nucleotides long. The contact size, d, represents the number of consecutive nucleotides required to maintain all contacts with the protein and is less than or equal to the occluded site size of the protein. In this model we assume that the protein utilizes its full contact size even when bound to the ends of the DNA (i.e., no dangling protein) (22). The NS3h monomer is initially bound i translocation steps away from the 5’-end, with concentration, Ii. The number of translocation steps, i, is constrained (1 ≤ i ≤ n), where n is the maximum number of translocation steps needed for a translocase bound initially at the 3’ end to move to the 5’ end of a DNA that is L nucleotides long. We assume that the binding of NS3h to the single-stranded DNA is random, but uniform, so that there is an equal probability of binding to any position along the single-stranded DNA with the exception of the ends of the DNA, which have a different probability of binding.
Upon addition of ATP the NS3h monomer moves with directional bias (from 3’ to -5’) along the DNA via a series of repeated rate-limiting translocation steps each associated with the same rate constant, kt. The rate constant for protein dissociation during translocation is kd. The finite processivity of translocation along the single-stranded DNA is thus defined as . Between each rate-limiting translocation step the protein moves m nucleotides and the product mkt is the macroscopic translocation rate and has units of nt/s. Inclusion of a protein trap prevents any free protein that dissociates during translocation from rebinding to the DNA (22, 25, 26). When the protein reaches the 5’-end of the DNA (I0 in Scheme 1) it dissociates from the DNA through a two-step process with associated rate constants kc and kend.
We note that, in general, kt represents the rate constant for the rate-limiting step that occurs within each repeated translocation cycle and does not necessarily correspond to the rate constant for physical movement of the protein along the DNA (22). Similarly, the average number of nucleotides translocated between two successive rate-limiting steps, defined as the translocation “kinetic step-size” (m), might be larger than the smallest length of DNA traversed during a single physical movement of the enzyme along the DNA.
Based on Scheme 1, the expressions in equation (1) can be derived (22, 25) for the time-dependent accumulation of protein at the 5’ end of the DNA.
| (1) |
In these equations, L−1 is the inverse Laplace transform operator, s is the Laplace variable, kt, kdkc kend and n are as defined above. The variable A is a scalar factor incorporating both the initial concentration of protein bound to the single-stranded DNA and the fluorescence signal change associated with protein being bound at the 5’ end of the DNA (in the I0 state in Scheme 1). The variable B is the ratio of the fluorescence signal change associated with a protein completing the second step of the two step dissociation process to the fluorescence signal change associated with the protein being bound at the 5’ end of the DNA. The variable r is the ratio of the initial probability (before the addition of ATP) of the enzyme being bound to any other position (other than the 3’ or 5’ ends) on the single-stranded DNA to the initial probability (before the addition of ATP) of the enzyme being bound to 5’ end of the DNA (22). The variable rn is the ratio of the initial probability (before the addition of ATP) of the enzyme being bound to the 3’ end of the single-stranded DNA to the initial probability (before the addition of ATP) of the enzyme being bound to 5’ end of the DNA. As shown in Equation 2, we can relate n to the kinetic step size of translocation (m) and the interaction size of the enzyme on the DNA (d) (21, 22, 25, 26).
| (2) |
Dissociation of NS3h from internal regions of ssDNA
Previous studies of the single-stranded DNA translocation activity of several bacterial helicases has relied upon independent measurements of the rate of dissociation from internal regions of the DNA (the kd variable in Scheme 1 and equation (1)) as a constraint in the NLLS analysis of the kinetic time courses (22, 25, 26, 46). In an attempt to mimic this procedure, we monitored the dissociation kinetics of NS3h through changes in the intrinsic tryptophan fluorescence of the enzyme following it's dissociation from poly(dT) and subsequent binding to heparin. NS3h (50 nM) was pre-bound to poly (dT) (5 μM nucleotide) in assay buffer, and dissociation was initiated by mixing with buffer containing 5mM ATP, and 10 mM Mg(OAc)2, and various concentrations of heparin. The observed time courses (shown in Figure 7) are clearly biphasic and well described by a simple two-exponential fits (equation 3). The results of linear least squares fitting of the data in Figure 7 to equation 3 are shown in Table 1.
| (3) |
The analysis of these time courses indicates that no further changes in the kinetics of NS3h translocation, dissociation and heparin binding occur beyond a concentration of 2 mg/ml heparin. This is consistent with our experimental results indicating that a heparin concentration of 2 mg/ml was sufficient to trap free NS3h under these solution conditions (Supplemental Figure 5). However, the biphasic nature of all of the time courses in Figure 7 (including those obtained in the absence of heparin) and the observation that the associated rate constants (k1,obs and k2,obs in Table 1) decrease with increasing heparin concentration make interpretation of the traces difficult. The second phase may represent photobleaching of NS3h over the time frame of the experiment. A control experiment was performed in the absence of ATP, and the decrease in fluorescence was similar to that observed in the presence of ATP (supplemental figure 3). The observation of a decrease in fluorescence in the absence of ATP indicates that the second phase is not likely related to translocation and may indeed be photobleaching For this reason, we have analyzed the data in two ways. The value of kd was allowed to float in the NLLS analysis and the value was fixed according to the first observed rate constant obtained in Figure 7.
Figure 7.
NS3h translocation along poly(dT) monitored by stopped-flow fluorescence spectroscopy of the intrinsic tryptophan fluorescence of NS3h for short (A) and long (B) timescales. NS3h (50 nM) was pre-bound to poly (dT) (5 uM nucleotide) in assay buffer, and translocation was initiated by mixing with buffer containing ATP (5 mM), Mg(OAc)2 (10 mM),and various concentrations of heparin. Fluorescence time courses are shown with final heparin concentrations of 0 mg/ml ( ), 2 mg/ml (
), 2 mg/ml ( ), 4 mg/ml (
), 4 mg/ml ( ), and 6 mg/ml (
), and 6 mg/ml ( ). (C) NS3h (100 nM) was pre-bound to 37:22mer (200 nM) in assay buffer, and translocation was initiated by mixing with buffer containing ATP (5 mM), Mg(OAc)2 (10 mM),and heparin (4 mg/ml). Fluorescence data was fit to the sum of two exponentials resulting in a rate constant of 2.0 ± 0.01 s−1 for the first exponential phase and 0.029 ± 0.006 s−1 for the second exponential phase of the progress curve.
). (C) NS3h (100 nM) was pre-bound to 37:22mer (200 nM) in assay buffer, and translocation was initiated by mixing with buffer containing ATP (5 mM), Mg(OAc)2 (10 mM),and heparin (4 mg/ml). Fluorescence data was fit to the sum of two exponentials resulting in a rate constant of 2.0 ± 0.01 s−1 for the first exponential phase and 0.029 ± 0.006 s−1 for the second exponential phase of the progress curve.
Table 1.
Linear least squares fitting of the data in Figure 7.
| [Heparin] (mg/ml) | k1,obs (s−1) | k2,obs (s−1) |
|---|---|---|
| 0 | 0.54 ± 0.04 | 0.038 ± 0.003 |
| 2 | 0.41 ± 0.05 | 0.026 ± 0.004 |
| 4 | 0.42 ± 0.03 | 0.018 ± 0.002 |
| 6 | 0.40 ± 0.04 | 0.022 ± 0.003 |
Mechanism of single-stranded DNA translocation by NS3h
Estimates of the kinetic parameters in Scheme 1 were obtained through the global nonlinear least squares analysis of the time courses in Figure 6 using Equation 1. In this analysis, kt, kd, kc, kend, m, d, B, r and rn were floated as global parameters (i.e., constrained to be the same for all single-stranded DNA lengths) whereas the A parameters were allowed to float independently for each single-stranded DNA length. The best fit parameters obtained from global NLLS analysis of time courses in Figure 6 are kt= 28(±4) steps s−1, kd = 0.2(±0.06) s−1, kc= 6 (±1) s−1, kend = 1.8(±0.1) s−1, and m = 1.7(±0.2) nt (summarized in table 2). For comparison, rate constants obtained when the kd value was fixed are reported in Supplemental Table 2. The macroscopic rate constant for translocation (m*kt) was determined to be 43 nt s−1 when the value for kd was allowed to float and 46 nt s−1 when the value was fixed.
Table 2.
The best fit parameters obtained from global NLLS analysis of data in Figure 6.
| Scheme 1 | Scheme 2 | ||
|---|---|---|---|
| kt (steps/s) | 27 ± 3 | kt (steps/s) | 30 ± 6 |
| kd (s−1) | 0.2 ± 0.06 | kd (s−1) | 0.20 ± 0.06 |
| kc (s−1) | 6 ± 1 | ki (s−1) | 6 ± 1 |
| kend (s−1) | 1.8 ± 0.1 | kend (s−1) | 1.8 ± 0.1 |
| m (nt/step) | 1.7 ± 0.2 | m (nt/step) | 1.4 ± 0.3 |
| d (nt) | 21 ± 4 | d (nt) | 19 ± 4 |
| r | 0.028 ± 0.004 | r | 0.024 ± 0.005 |
| rn | 0.22 ± 0.05 | rn | 0.19 ± 0.04 |
| B | 8.3 ± 1.3 | C | 0.30 ± 0.08 |
| m*kt (nt/s) | 46 ± 5 | m*kt (nt/s) | 46 ± 5 |
| m/(1-P) (nt) | 230 ± 60 | m/(1-P) (nt) | 230 ± 80 |
| Variance | 4.39 × 10−10 | Variance | 4.39 × 10−10 |
An equally probable representation of the kinetic mechanism for single-stranded DNA translocation by NS3h is shown in Scheme 2 and the associated equation for the fluorescence is shown in equation 4.
| (4) |
In Scheme 2, the NS3h monomer is initially bound in a state that is not competent for processive DNA translocation (the Ii* concentrations in Scheme 2). The variable ki in this scheme and equation 4 is the rate constant for the translocation initiation process that occurs after the binding of ATP; the variables kt, kd, kend, m, d, r and rn in Scheme 2 and equation 4 are as defined for Scheme 1 and equation 1. The variable C is the ratio of the fluorescence signal change associated with an enzyme bound at the 5’ end of the DNA in the pre-initiation state (the Io* concentration in Scheme 2) to the fluorescence signal change associated with an enzyme bound at the 5’ end in the post-initiation state (the I0 concentration in Scheme 2). The best fit parameters obtained from global NLLS analysis of time courses in Figure 6 according to Scheme 2 (equation 4) are kt= 30(±6) steps s−1, kd = 0.2(±0.06) s−1, kc= 6 (±1) s−1, kend = 1.8(±0.1) s−1 , and m = 1.4(±0.3) nt (summarized in Table 2). The fits of the data in Figure 6 using equation 4 overlay perfectly with the fits obtained using equation 1 (as indicated by the identical variance of the fit in Table 2).
It is worth noting that correlation exists between some sets of parameters in these equations; the parameters kt, m, r and rn are especially highly correlated with each other, for example. Therefore, coupled variations in these parameters can result in nearly equally good fits of the data (see Supplemental Table 1). We would like to emphasize, however, that the estimate of the macroscopic translocation rate (m*kt) is well constrained by the data, regardless of variations in kt or m. As shown in supplemental material, although reasonable fits of the time courses, as judged by their variances, are obtained when kt is constrained to values in the range from 27 to 270 steps/s, the estimate of the macroscopic translocation rate remains similar. Similarly, when the kd value for dissociation of NS3h from poly dT is fixed based on the rate of the first exponential from data in Figure 7, the macroscopic rate constant for translocation remains similar to that obtained when allowing the kd value to float (Supplemental Table 2). Taken together, we believe that the macroscopic translocation rate is more accurately constrained by our analysis than the associated microscopic translocation rate. Similarly, we believe that the estimate of the macroscopic translocation rate is more accurately constrained than the estimates of the binding parameters r and rn.
Translocation experiments were conducted with Cy3-labeled oligonucleotides in order to compare results obtained with fluorescein-labeled oligonucleotides (supplemental Figure 4). The estimate of d is larger for the Cy3 substrates and the estimate of m*kt is smaller for the Cy3 substrates (supplemental table 3). Similar trends in the variation of the estimates of d and m*kt between F- and Cy3-labeled DNA were observed previously in the case of UvrD and were attributed to a difference in the physical distances over which UvrD could interact with these fluorophores and subsequently affect their fluorescence (20). Our results suggest that a similar difference in interaction distance between NS3h and these same fluorophores also occurs.
Dissociation of NS3h from DNA unwinding substrate
The lack of product formation during unwinding by monomeric NS3h under single cycle conditions is surprising in light of the relatively fast rate of translocation on ssDNA (~ 46 nt/s). The unwinding rate measured in the presence of excess concentration of NS3h relative to DNA was 0.61 s−1 for the 22 bp substrate (Figure 1). The final 8-10 base pairs melt spontaneously (38), so we can assume that ~12 bp are melted by the action of the helicase, leading to a rate for unwinding of 7.3 bp/s. The movement of NS3h along DNA clearly slows upon encountering duplex DNA.
The dissociation rate of NS3h from the ss/dsDNA substrate was measured by using stopped-flow fluorescence spectroscopy under conditions that were identical to those used for measuring translocation. NS3h (100 nM) was incubated with the 37:22mer (200 nM) followed by rapid mixing with ATP, Mg2+, and heparin, resulting in a dissociation rate constant of 2.0 ± 0.01 s−1 (Figure 7C). This dissociation rate constant is ~5-fold faster than dissociation from ssDNA (compare to Figure 7A). Therefore, when NS3h encounters duplex DNA, it dissociates faster than when translocating on ssDNA. The combination of slower movement through the duplex and faster dissociation leads to relatively poor DNA unwinding activity by NS3h when compared to its translocase activity.
DISCUSSION
The mechanism of processive DNA unwinding by helicases has been linked to the mechanism of translocation on ssDNA (16, 22). Mechanisms have been proposed in which melting of a base pair occurs, followed by translocation of one nucleotide in a process that is coupled to ATP binding and hydrolysis. The steps for melting the dsDNA and translocation on ssDNA have been proposed to be separable in some cases (43). For other helicases such as Dda, dsDNA unwinding has been proposed to be a consequence of translocation on ssDNA (13, 17) whereby movement of the enzyme along the DNA forces apart the base pairs. It is clear, however, that some helicases can readily translocate on ssDNA, yet are unable to unwind duplexes under single cycle conditions (21, 25). Hence, the ability to translocate on ssDNA is not necessarily sufficient for DNA unwinding.
Protein displacement has been suggested as an important biological role played by helicases. During homologous recombination in Saccharomyces cerevisiae, Rad51 protein serves as a recombinase by forming filaments on ssDNA (47, 48). Srs2 is a superfamily 1 helicase that disassembles Rad51 nucleoprotein filaments, thereby suppressing homologous recombination (49, 50). The physical interaction between Srs2 and Rad51 triggers ATP hydrolysis within the Rad51 filament, causing Rad51 to dissociate from DNA (51). However, an ATPase-deficient form of Srs2 was unable to stimulate disassembly of Rad51 filaments, indicating that translocase activity of Srs2 also plays a role in removing the protein filament. Understanding the relationships between translocation, DNA unwinding, and protein displacement is necessary to define the biochemical activities of many helicases.
NS3 (and/or NS3 helicase domain) from the Hepatitis C virus has become one of the most studied SF2 helicases. In addition to its medical importance related to viral infection and liver disease, NS3 is considered a model system for this group of enzymes. Translocation and melting have been proposed to be linked, but occur in separate steps in a ‘spring-loaded’ mechanism, based on stepping measurements made using single molecule FRET to study DNA unwinding (36). Here we have investigated the relationship between translocation, DNA unwinding, and protein displacement by examining each reaction under identical conditions.
Full-length NS3 contains a helicase domain and a protease domain which can be expressed and studied separately. Each form of the enzyme was studied under identical conditions in this report. Little or no unwinding is observed for each form of the enzyme when the DNA substrate concentration is in excess of the enzyme concentration under single cycle conditions (Figure 2). The lack of product does not mean that the monomeric enzyme does not unwind DNA. When one molecule of enzyme binds to the DNA substrate, the helicase dissociates prior to unwinding a sufficient portion of the 22 bp substrate to observe ssDNA under single cycle conditions. Under identical conditions (substrate concentration > enzyme concentration), NS3h clearly translocates on ssDNA (Figure 6). The number of nucleotides translocated in a single binding event is around 230 nt. The fact that only 22 bp are not unwound in a single cycle indicates that unwinding of dsDNA alters the activity of NS3h significantly. The rate of DNA unwinding by NS3h is reduced compared to the rate of translocation on ssDNA. The rate of dissociation of NS3h from the duplex substrate was 5-fold faster than dissociation from ssDNA. DNA unwinding by NS3h has been shown to be sensitive to the stability of the base pairs within the duplex (52). Movement of the enzyme is therefore impeded by duplex DNA and the results here are consistent with this interpretation.
Under conditions in which enzyme concentration exceeds DNA substrate concentration, NS3h unwinds the 22bp substrate (Figure 1). Multiple molecules of NS3h bound to the same DNA substrate molecule can increase product for DNA unwinding through functional cooperativity (38). Single-stranded DNA binding proteins can also function with NS3h to increase DNA unwinding activity, possibly by interacting directly with the helicase and by preventing reannealing of the DNA after helicase-catalyzed unwinding (39). NS3h clearly requires other molecules for optimal DNA unwinding activity. The reason for this requirement may relate to the affinity of NS3h for DNA during the ATP hydrolysis cycle. A “weak-state” of interaction has been reported for the ATP-bound form of NS3h (53). The role served by SSB or additional helicase molecules may be as processivity factors that hold NS3h onto the DNA during the weak binding state of the cycle, thereby enhancing the opportunity for overcoming the barrier presented by dsDNA.
Determining the relationship between ssDNA translocation and DNA unwinding by NS3h is made difficult by the fact that NS3h does not readily unwind DNA under the conditions in which translocation can be measured. If NS3h-catalyzed DNA unwinding were observed under these conditions, then kinetic parameters from the two methods could be directly compared with confidence. However, the kinetic parameters for translocation can be compared to kinetic parameters for DNA unwinding obtained from experiments under excess enzyme conditions. The kinetic step size obtained for translocation of NS3h on ssDNA was 1.4 – 1.7 bp per step (Table 2). We note that this microscopic parameter is less well-constrained than macroscopic parameters such as the rate of translocation. However, it is worth discussing this value in the context of measurements of step size provided by other labs.
Such a relatively small kinetic step size implies that the physical step size for NS3h is likely to be 1 or 2 base pairs (34). Larger kinetic step sizes have been reported for DNA and RNA unwinding. A kinetic step size of 9 bp was measured for NS3h under excess enzyme conditions (38). RNA unwinding studies resulted in a kinetic step-size of ~ 18 bp for full-length NS3 (33). The larger kinetic step sizes are likely made up of smaller, sub-steps. Single-molecule approaches have provided measurements of the physical step size for full-length NS3 of 11 bp for RNA unwinding with smaller substeps of 2-3 bp (35). Single-molecule FRET experiments were interpreted in terms of a physical step size of ~ 3 bp, which was made up of smaller 1 bp sub-steps (36). The smaller steps measured by single molecule measurements indicate that one ATP hydrolysis event per NS3 molecule is likely to lead to movement by one bp.
Most recently, Serebrov et al. demonstrated that kinetic step size (16bp/step) was independent of RNA duplex stability and composition but is dependent on salt concentration (54). Unwinding was enhanced under low salt conditions by monomeric NS3 and step size was decreased to 11 bp/step. The “super-steps” (11 and 18 bp) observed in RNA unwinding were suggested to be caused by delayed product release of separated RNA product from domain II of NS3 (54). The kinetic step size of NS3h, 1.4 – 1.7 nt, reported here under monomeric conditions can be readily interpreted to indicate a movement of one or two base pairs per ATP binding and hydrolysis event, which is consistent with the results from single molecule FRET studies. The fact that DNA unwinding is not occurring during the translocation assays means that product release of the displaced strand is not part of the mechanism and may explain the lack of “super-steps” observed in the translocase assay. Direct measurements of ATPase activity are needed to further test the idea that NS3h moves by one base per ATP hydrolysis event (16, 21, 23).
Attempts to examine translocation using fluorescently-labeled oligonucleotides did not succeed for full-length NS3 (supplemental figure 1). The lack of clearly defined changes in fluorescence as a function of oligonucleotide length for NS3 might be a result of the oligomeric nature of the enzyme under these conditions. For example, oligomeric NS3 might exist in a form that binds to multiple sites along the single-stranded DNA, essentially coating the DNA. This would lead to the fluorescent probe being bound by NS3 at the start of the reaction, regardless of the length of the oligonucleotide. Much longer oligonucleotides might be needed to measure translocase activity of oligomeric NS3, or an alternative method needs to be developed that examines each individual binding site along the DNA. ATPase activity has been measured as a function of ssDNA length for several helicases to infer translocation mechanisms, and such methods may apply for full-length NS3 (23).
Displacement of streptavidin is observed for NS3, but not for NS3h, and only when NS3 concentration is in excess of substrate concentration (Figures 3 and 4). Others have reported streptavidin displacement by NS3h (55), however we were unable to observe this activity under conditions described here. The lack of displacement by NS3h is surprising, especially in light of the demonstrable translocase activity. This result illustrates that translocation per se does not provide sufficient force to dislodge streptavidin from the biotin-labeled oligonucleotides, at least under conditions examined here. Force-production might require specific interactions between helicase and DNA that are not present in NS3h. The x-ray crystal structure of NS3h bound to DNA indicates that the majority of interactions are electrostatic in nature, although some stacking interactions between nucleobase and amino acids are also observed (40). Full-length NS3 binds more tightly to DNA than NS3h, which might allow greater force production during translocation due to additional contacts made with nucleic acid through the protease domain of NS3 (32, 54).
Interestingly, our results also suggest a preference for NS3h to bind to the ends of the single-stranded DNA, rather than to the middle (r << 1 in Table 1). This is in contrast to what has been observed with several bacterial helicases (UvrD, PcrA, Rep) and warrants further investigation. This preference for binding to the ends of the single-stranded DNA may also be responsible for the apparent inflation of the estimate of the interaction size of NS3h on the single-stranded DNA (d ~ 20 nt in Table 1).
The experiments reported here indicate that NS3h translocates 46 ± 5 nt/s with a step size of 1.4-1.7± 5 nt/step and a processivity of 230 ± 60 nt. From these results one can predict that NS3h should unwind short duplex DNA and displace proteins bound to DNA (streptavidin) under identical conditions. Interestingly NS3h was unable to unwind the 22 bp duplex substrate and could not displace streptavidin from biotin labeled DNA under the conditions in which translocation was measured. Therefore, optimal DNA unwinding and protein displacement by NS3h requires more than simply translocation. Interaction with other proteins such as single-stranded binding proteins (39) or function with additional molecules of NS3h (38) can result in enhanced helicase activity for this ssDNA translocase.
Supplementary Material
Footnotes
This work was supported by NIH research grants R24 GM080599 (K.D.R.) and R01 GM089001 (K.D.R and C.E.C.).
SUPPORTING INFORMATION AVAILABLE
Supplementary file provides data on size exclusion chromatography of NS3h in the presence of ssDNA, NS3 translocation on 5’-F-(dT)L, NS3 translocation on Poly(dT)/M13, NS3h translocation on Cy3-labeled oligos, NS3h translocation in the presence of different concentrations of heparin, and NS3h translocation on biotinylated oligo (3’T58BiodTG5’) in the presence and absence of streptavidin. The supporting information is available for free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 3.Patel SS, Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 4.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya AE, Koonin EV. Helicases; aminoacid sequence comparisions and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 6.Maluf NK, Fischer CJ, Lohman TM. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 7.Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 9.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 10.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 11.Eggleston AK, O'Neill TE, Bradbury EM, Kowalczykowski SC. Unwinding of nucleosomal DNA by a DNA helicase. J. Biol. Chem. 1995;270:2024–2031. doi: 10.1074/jbc.270.5.2024. [DOI] [PubMed] [Google Scholar]
- 12.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 13.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 14.Morris PD, Byrd AK, Tackett AJ, Cameron CE, Tanega P, Ott R, Fanning E, Raney KD. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5'-biotinylated oligonucleotides but not from 3'-biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 15.van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 16.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 17.Tackett AJ, Morris PD, Dennis GE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 18.Walstrom KM, Dozono JM, Robic S, von Hippel PH. Kinetics of the RNA–DNA Helicase Activity of Escherichia coli Transcription Termination Factor Rho. 1. Characterization and Analysis of the Reaction. Biochemistry. 1997;36:7980–7992. doi: 10.1021/bi963179s. [DOI] [PubMed] [Google Scholar]
- 19.Walstrom KM, Dozono JM, von Hippel PH. Kinetics of the RNA–DNA Helicase Activity of Escherichia coli Transcription Termination Factor Rho. 2. Processivity, ATP Consumption, and RNA Binding. Biochemistry. 1997;36:7993–8004. doi: 10.1021/bi963180r. [DOI] [PubMed] [Google Scholar]
- 20.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 22.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maluf NK, Ali JA, Lohman TM. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 25.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 26.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J. Bacteriol. 2007;189:4502–4509. doi: 10.1128/JB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veaute X, Delmas S, Selva M, Jeusset J, Cam EL, Matic I, Fabre F, Petit M-A. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia Coli. EMBO. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai CL, Chi WK, Chen DS, Hwang LH. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 31.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 32.Sikora B, Chen Y, Lichti CF, Harrison MK, Jennings TA, Tang Y, Tackett AJ, Jordan JB, Sakon J, Cameron CE, Raney KD. Hepatitis C virus NS3 helicase forms oligomeric structures that exhibit optimal DNA unwinding activity in vitro. J. Biol. Chem. 2008;283:11516–11525. doi: 10.1074/jbc.M708125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 34.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: Quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J. Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 35.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 38.Levin MK, Wang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopal V, Patel SS. Single strand binding proteins increase the processivity of DNA unwinding by the hepatitis C virus helicase. J. Mol. Biol. 2008;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J. Biol. Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 41.Williams DJ, Hall KB. Monte Carlo application to thermal and chemical denaturation experiments of nucleic acids and proteins. Methods Enzymol. 2000;321:330–352. doi: 10.1016/s0076-6879(00)21201-9. [DOI] [PubMed] [Google Scholar]
- 42.Porter DJ, Preugschat F. Strand-separating activity of Hepatitis C Virus helicase in the absence of ATP. Biochemistry. 2000;39:5166–5173. doi: 10.1021/bi992384f. [DOI] [PubMed] [Google Scholar]
- 43.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter DJT, Short SA, Hanlon MH, Preugschat F, Wilson JE, Willard DH, Jr., Consler TG. Product release is the major Contributor to kcat for the Hepatitis C Virus Helicase-catalyzed Strand Separation of Short Duplex DNA. J. Biol. Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 45.Raney KD, Benkovic SJ. Bacteriophage T4 Dda helicase translocates in a unidirectional fashion on single-stranded DNA. J. Biol. Chem. 1995;270:22236–22242. doi: 10.1074/jbc.270.38.22236. [DOI] [PubMed] [Google Scholar]
- 46.Fischer CJ, Lohman TM. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J. Mol. Biol. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol.Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 48.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 49.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 50.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 51.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 Disassembles Rad51 Filaments by a Protein-Protein Interaction Triggering ATP Turnover and Dissociation of Rad51 from DNA. Molecular Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donmez I, Rajagopal V, Jeong Y-J, Patel S. Nucleic acid unwinding by Hepatitis C virus and bacteriophage T7 helicases is sensitive to base pair stability. 2007. pp. 21116–21123. [DOI] [PubMed]
- 53.Levin MK, Gurjar M, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 54.Serebrov V, Beran RK, Pyle AM. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J. Biol. Chem. 2008;284:2512–22521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.