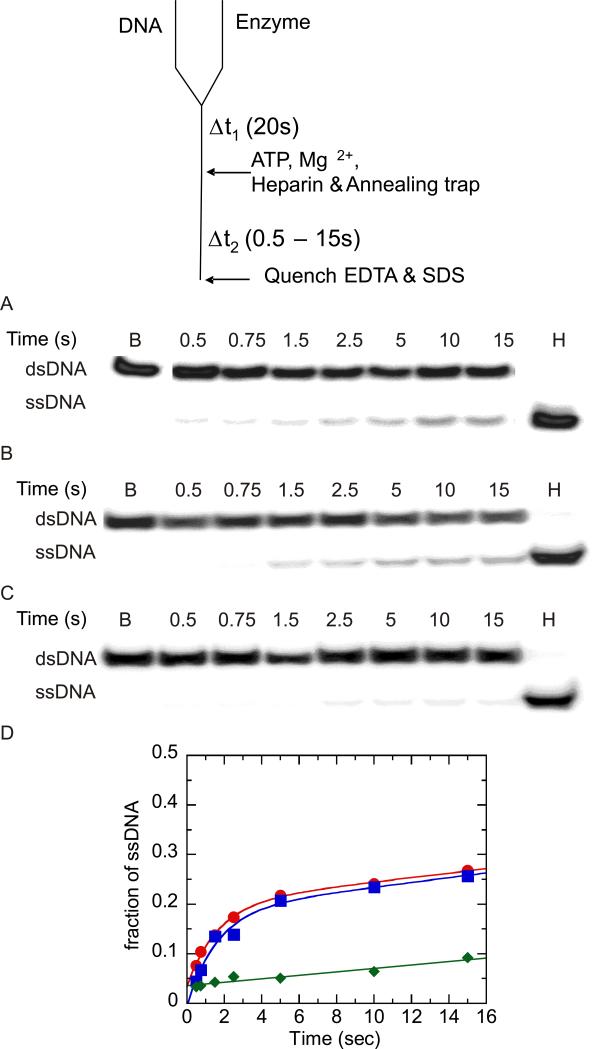
Figure 1.
Unwinding of 37:22-mer partial duplex by NS3 and NS3h in the presence of heparin (used as a protein trap) under conditions of enzyme concentration in excess of DNA substrate concentration. A “double-mixing” experimental protocol was performed as described in the text. Unwinding products were resolved on 20% polyacrylamide gel and visualized using a PhosphorImager and quantitated by using ImageQuant software. (A) Representative gel image showing unwinding of 2 nM partial duplex by 500 nM NS3. The blank sample (B) shows the partial duplex DNA substrate prior to unwinding, and sample (H) shows the DNA after heating to 95 °C for ten min. (B) Representative gel image showing unwinding of 2 nM partial duplex by 500 nM NS3h. (C) To determine the efficiency of heparin as a protein trap, the partial duplex substrate (2 nM) was mixed with heparin (4 mg/ml) prior to initiating the reaction upon mixing with NS3h (500 nM). Little or no unwinding was observed. (D) Fraction of product formation for unwinding of 2 nM partial duplex by 500 nM NS3 ( ), 500 nM NS3h (
), 500 nM NS3h ( ), and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. Enzyme-catalyzed unwinding was fit to a single exponential followed by a steady state rate resulting in amplitudes of 0.19 ± .03 and 0.17 ± .01 nM, rate constants of 0.62 ± 0.3 and 0.61 ± 0.1 s−1, and steady state rates of 0.005 ± .001 and 0.004 ± 0.001 nM/s for NS3 and NS3h, respectively. The control experiment in which heparin was added to the DNA substrate prior to addition of NS3h was fit to a linear equation resulting in a rate of 0.004 ± 0.001 nM/s.(
), and 500 nM NS3h under conditions where heparin and partial duplex DNA were mixed together prior to initiation of the reaction by addition of enzyme. Enzyme-catalyzed unwinding was fit to a single exponential followed by a steady state rate resulting in amplitudes of 0.19 ± .03 and 0.17 ± .01 nM, rate constants of 0.62 ± 0.3 and 0.61 ± 0.1 s−1, and steady state rates of 0.005 ± .001 and 0.004 ± 0.001 nM/s for NS3 and NS3h, respectively. The control experiment in which heparin was added to the DNA substrate prior to addition of NS3h was fit to a linear equation resulting in a rate of 0.004 ± 0.001 nM/s.( ).
).