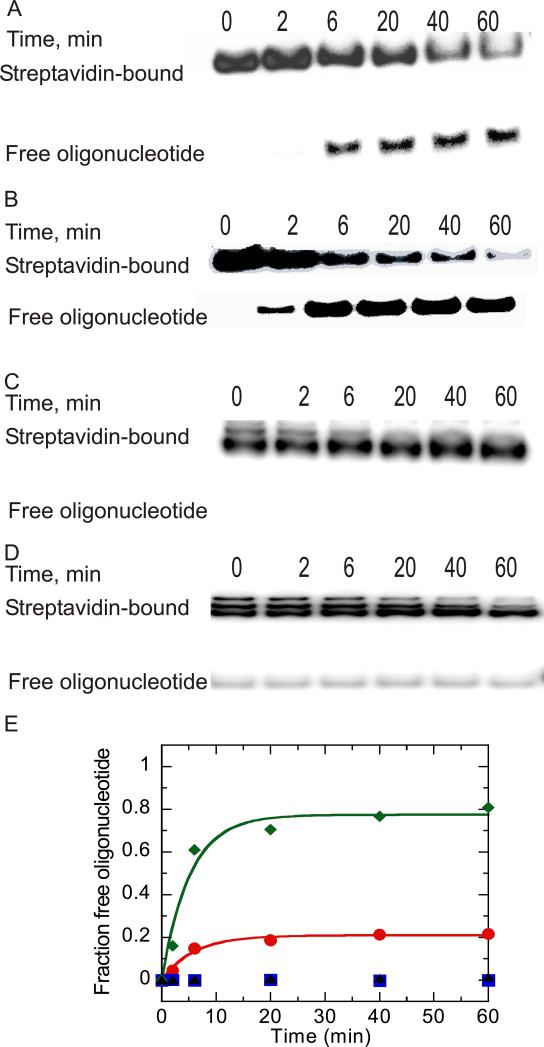
Figure 3.
NS3 catalyzed streptavidin displacement from 5’-bio-30mer and 5’-bio-60mer oligonucleotides. (A) NS3 (1 μM) was incubated with 5’-bio-30mer (10 nM) in reaction buffer. Samples were removed at varying times and added to a quencher solution (0.6% SDS, 200 mM EDTA, 0.08% Xylene cyanol, 0.08% bromophenol blue, and 10% glycerol) followed by separation of streptavidin-bound oligonucleotide from free oligonucleotide on a 15% native polyacrylamide gel. (B) NS3 (1 μM) was incubated with 5’-bio-60mer 10 nM in reaction buffer and treated as described for panel A.. (C) NS3 (100 nM) was incubated with 5’-bio-30mer 200 nM under identical reaction conditions as described for panel A, and samples were analyzed by 15% native polyacrylamide gel electrophoresis. (D) NS3 (100 nM) was incubated with 5’-bio-60mer (200 nM) under identical conditions as described for panel A. (E) Streptavidin displacement from different lengths of biotin oligonucleotides catalyzed by NS3. Results were obtained by quantitation of gels in panels A-D. Streptavidin displacement over time is shown in the presence of NS3 (1 μM) incubated with 10 nM 5’-bio-30mer ( ),NS3 incubated with 10 nM 5’-bio-60mer (
),NS3 incubated with 10 nM 5’-bio-60mer ( ), NS3 (100 nM) incubated with 200 nM 5’-bio-30mer (
), NS3 (100 nM) incubated with 200 nM 5’-bio-30mer ( ), and NS3 (100 nM) incubated with 200 nM 5’-bio-60mer (▲).
), and NS3 (100 nM) incubated with 200 nM 5’-bio-60mer (▲).