Abstract
The desideratum of semantic interoperability has been intensively discussed in medical informatics circles in recent years. Originally, experts assumed that this issue could be sufficiently addressed by insisting simply on the application of shared clinical terminologies or clinical information models. However, the use of the term ‘ontology’ has been steadily increasing more recently. We discuss criteria for distinguishing clinical ontologies from clinical terminologies and information models. Then, we briefly present the role clinical ontologies play in two multicentric research projects. Finally, we discuss the interactions between these different kinds of knowledge representation artifacts and the stakeholders involved in developing interoperational real-world clinical applications. We provide ontology engineering examples from two EU-funded projects.
Keywords: Clinical Ontologies, Formal Ontologies, Knowledge Representation
1 Introduction
The desideratum of semantic interoperability has been intensively discussed in medical informatics circles over the past decade [Rossi-Mori 98, Ingenerf 01,Garde 07]. Consider, as examples, the evolution of the Unified Medical Language System (UMLS) [UMLS 07], of the HL7 Common Document Architecture (CDA) [Dolin 06], of openEHR [Kalra 05], or of the SNOMED CT [SNOMED CT 09].
Originally the issue of semantic interoperability was supposed to be addressed mainly by applying clinical terminologies. However, we have seen steady growth in usage of the term ‘ontology’ more recently with its occurrences having increased 17-fold in PubMed abstracts since the year 2000. Also, initiatives such as the Open Biomedical Ontologies (OBO) repository [OBO-Foundry 08], which is constantly growing in size and coverage, and which has embraced the goal of covering the entirety of the biomedical domain with a suite of high-quality ontologies through the OBO Foundry [Smith et al. 07], can be seen as further signs of the increasing focus on adequate semantic interoperability.
Some researchers have argued that the movement towards ontologies and away from pure terminologies constitutes no real scientific advance. And indeed, some good reason can be found to – at least partially – cast doubt on some of the claims made on behalf of ontologies: Too many recent publications, calls for research proposals and project descriptions have proven themselves to rest on insupportable expectations. Thus, it is easy to understand why some researchers are tempted to see in the term ‘ontology’ just yet another flashy catchword.
In the following article, we are contenting ourselves with the attempt to clearly capture the difference between terminology on one side and ontology on the other. But since the problem already starts with the fact that neither of those two terms owns a generally accepted definition, we have to adhere to the following two often cited definitions for our deliberations:
Terminology =def. A set of terms representing the system of concepts of a particular subject field. [ISO 00]
Ontology =def. The study of what there is [Quine 48]. Formal ontologies are theories that attempt to give precise representations of the types of entities in reality, of their properties and of the relations between them, using axioms and definitions that support algorithmic reasoning.
2 Delimiting the Concept of Ontology
2.1 Terminologies
Terminologies are term-centered and relate the various senses or meanings of linguistic entities with each other, for example by recording the synonymy between terms like “Nephroblastoma” and “Wilms’ Tumor”. In terminologies, classes of (quasi-) synonymous terms are commonly referred to as ‘concepts’, and in fact, in many communities this term actually stands for ‘meaning’. But unfortunately this particular usage of the term is oftentimes also associated with a variety of other confusing usages, for example, in that concepts are simultaneously also the referents of such terms. In many terminology systems (often also called ‘thesauri’ or ‘semantic lexica’), concepts are furthermore related only by highly informal semantic relations often defined (at best) by using vague natural language predicates1 (e.g., “Treatment hasBroaderMeaningThan Surgery”).
In medical informatics research, this particular language-centered view characterizes the legacy of the UMLS [UMLS 09]. Yet, in spite of its well-known shortcomings, the UMLS has proven itself to constitute a highly robust and useful platform for the retrieval of data associated with terms drawn from a large array of heterogeneous clinical terminology systems.
2.2 Ontologies
Ontologies are intended to describe some given portions of reality. In building them we can rely on logically defined – and hence precise – formalisms which contain special symbols and constructs making it possible to describe reality without the need to depend on human language descriptions that are often very ambiguous. Therefore, it is just for the convenience of ontology developers and users to use English language terms for describing the ontology content in addition to the logical expressions (employed by software tools). Such descriptions allow the users to better understand the intended meaning of the classes and properties within an ontology. But for the underlying formalism, natural language expressions play no role at all. Hence, the actual constituent nodes of an ontology represent neither terms nor (language) concepts but rather types on the side of the entities in reality [Smith et al. 06].
Such types (often also referred to as ‘categories’, ‘kinds’, or ‘universals’) are the basic ontological building blocks and reflect a hierarchical order in the organization of reality (including the medically relevant reality, such as patients, pathogens, lesions, or surgical procedures). The main construction tenet for ontologies is the taxonomic principle to the effect that a type S is a subtype of another type T if and only if all instances of S are also instances of T (see Fig. 1).
Figure 1.
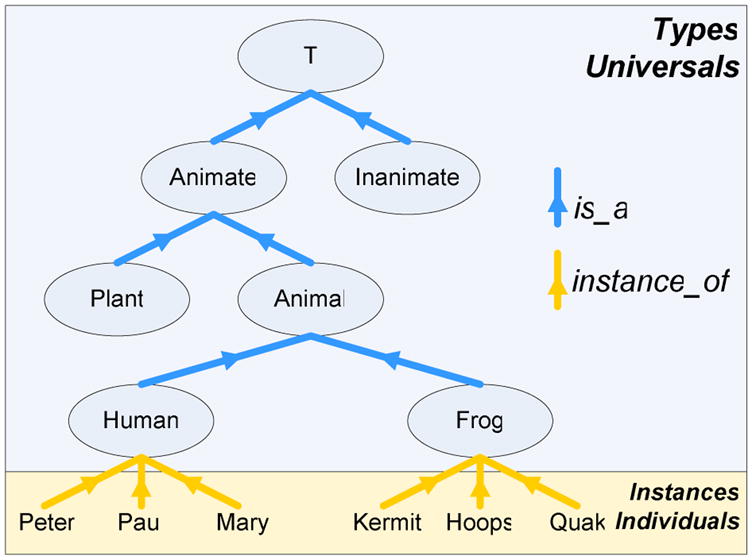
Simple subsumption hierarchy where types are in is_a relation if and only if all instances of the child classes are instances of the parent class.
The question of whether types do really exist – in addition to their instances in the real world – is still subject to major disputes among philosophers, as also are the criteria which could be used to determine what general terms should be accepted as representing types in reality. However, at any given stage in the development of science, a consensus core of textbook-scientific understanding is reached, and this core together with the repertoire of types which it contains, should – in our understanding – serve as starting point in developing any science-based ontology. Simple examples of statements belonging to this consensus core are that humans are mammals, that cells contain membranes, or that the retina contains photosensors.
However, phenomena of the sort which are of concern in medicine are elucidated not only by observation-based descriptions of nature involving representations of natural kinds of these sorts. In many times, they refer to entities reflecting prescriptive definitions or specifications made by human experts. Some medical examples are “appendectomy” defined as the “surgical procedure to remove the appendix”, “hepatitis” defined as an “inflammation of liver tissue”, or “hypertension” defined as “abnormally high arterial blood pressure” [Merriam-Webster 08].
The types in an ontology correspond to entities existing in the real world (also called ‘individuals’ or ‘instances’), and forming classes that extend these types. Classes themselves fall into two groups, namely those which are extensions of corresponding types, and those we call ‘defined classes’ [Smith et al. 06], for which there exists no corresponding type. Examples of the latter include the “class of diabetics living in Freiburg on some given day”, or the “class of appendectomies performed in a given hospital in some given month”.
2.3 Constraints on Ontology Structure and Content
Based on the proposed understanding of ontologies as representations of entity types and the relations between them, we can already rule out some of the common misconceptions that often obscure the sharp difference between ontologies, terminologies and other artifacts such as representations of contextual knowledge or clinical information models for data acquisition.
As a fundamental principle, all properties associated with any given type in any ontology must be true for all instances of this particular type. Thus, as an example, all instances of appendectomy are performed on some instance of appendix and all instances of water molecules contain oxygen and hydrogen. But in turn, this fact restricts the ability of any ontology to express assertions that are seemingly obvious: For instance, “hands have thumbs” or “aspirin alleviates headache” cannot be expressed unconditionally in an ontology, because there exist hands without thumbs (e.g., because of an accident or some congenital defect) and not all aspirin tablets are used to alleviate a headache (e.g., one can take aspirin for anticoagulation as well). In order to avoid such problems there has been a recent move to what is called canonical ontologies presenting idealized views on specific domains and basically allowing statements such as the ones above to be true ‘canonically’. The Foundational Model of Anatomy (FMA) presents such an idealized view for the description of the canonical human anatomy [Rosse et al. 03].
A related problem is the one of probabilistic assertions, which are of tremendous importance for everyday clinical reasoning, but that can currently not be expressed in any straightforward way in an ontology [Kayaalp 05]. For example, in the case that a prevalence of one percent is ascribed to lung cancer we are not asserting some property inhering in any particular instance of lung cancer. Rather, we are making a factual assertion about some given population. All assertions built into an ontology for representing the type “Lung Cancer” should apply ipso facto to all the instances of this particular type. However, the common consensus in many areas of science is precisely probabilistic in nature, and therefore describes results in terms of probabilistic states, processes, and events. This is especially the case in medicine for the assessment of risks (i.e., of particular conditions, or behavior, or therapies in relation to specific diseases): If we assume that there is a relation between the type “Arterial Hypertension” and the type “Stroke”, namely that the former is a risk factor for the latter, then there is a problem in explaining how this relation can be represented in formal ontologies following the principles described above.
A possible solution is to introduce probabilistic dispositions [Jansen 07] into the ontology, i.e., dispositions to do something under certain circumstances incorporating a certain probability. Such dispositions are related to the corresponding events by the relation of realization. They are special kinds of dependent entities in that they do not need to be realized in order to exist. The “Risk for Stroke”, for instance, could be represented in this way.
The just introduced fundamental constraints are corollaries of the fact that all assertions of relations between types in ontologies should follow the basic form of universal statements, i.e., “for all instances of type T there is some instance of type S such that…”. Of course, we could, consider both types and instances as two different ranges for our quantifiers. But without resorting to any “workarounds” or adaptations (for example along the lines of [Smith 05]), we would have to accept some type of higher-order logic in this case, which in turn could cause problems for machine reasoning systems, since such type of logics are known not to be computable in all circumstances with the current algorithms. In contrast, languages from the family of Description Logics [Baader et al. 03] are “well-behaved” in that they are known to be always computable, and are therefore frequently used in the development of ontologies.
2.4 Epistemological Classification Criteria
Classes are the basic building blocks for such clinical classification systems as the International Statistical Classification of Diseases (ICD) [WHO 09], which still provides our most significant support for semantic interoperation of clinical data, for the time being and on a global perspective.
It has repeatedly been observed that medical classification systems (even those claiming to classify entities in reality) should be distinguished from ontologies because they use classification criteria that are ‘non-ontological’ (i.e., not such as to represent the knowledge-independent reality on the side of the patient) but rather ‘epistemological’ (i.e., such as to represent the knowledge one has about these entities) [Bodenreider et al. 04]. Thus the current ICD makes a classificatory distinction between two different kinds of tuberculosis – those cases diagnosed by bacterial culture and those diagnosed by histology. Of course, a disease is not different in nature just because a different method is used in its diagnosis, because a classification creator has not established a class suitable for it, or because a physician lacks knowledge.
Another example can be found in the so-called residual classes (commonly identified by the numbers 8 and 9 as the fourth digit in the ICD code), that either refer to the classification system proper (“NEC – not elsewhere classified”) or to the state of knowledge of the coder (“NOS – not otherwise specified”).
Albeit such epistemological issues are non-ontological by nature, they are nevertheless crucial for medical documentation. Diagnostic statements tend to be error-prone, and vital decisions are very often based solely on brittle evidence – necessary or desirable information may simply be missing. Hence, a place must exist for encoding all information one actually holds in some practically available form, further including associated meta-data on how the information was acquired, one’s level of confidence in the validity of the information, and so forth.
However, an ontology is not the right place for information of this sort. General terms such as “unspecified tumor stage” or “infection of unknown origin” do not represent special kinds of tumor stage or of infection but rather express a certain state of knowledge. To include them in an ontology would reflect a confusion of “what is” with “what we know” about it. As said above, such contextual knowledge is important in clinical environments, but its proper expression requires some additional means of encoding and storage and should form part of the representation of the clinician’s knowledge of specific instances.
3 Clinical Ontologies in Practice
The questions we have addressed so far arise, to different degrees, in cases where domain ontologies – in the introduced sense – are projected to improve data acquisition, standardization, interoperation, as well as data analysis in the framework of both clinical research projects and routine clinical documentation.
Now, we briefly report on the experiences within the projects ACGT (Advancing Clinico-Genomic Trials on Cancer) [ACGT 09] and @neurIST (Integrated Biomedical Informatics for the Management of Cerebral Aneurysms) [Aneurist 09]. In both projects, customized ontologies are being developed to create a common basis for applications to semantically mediate between the various distinct software components within the projects.
The overall goal of ACGT and @neurIST is to create integrated information technology infrastructures by implementing common software platforms. The main intention is to improve the management of the respective diseases by creating an efficient processing and presentation environment that allows for the combination of existing knowledge with newly collected or generated data. The overall goal of the developed platforms is the integration of data from various sources and disciplines within the projects (e.g., clinical studies, genomic research and patient management). These data are currently highly fragmented and heterogeneous in regard to format and scale, and the way in which their content is collected and presented reflects disciplinary habits entrenched within the subdisciplines involved in each of the projects.
The ACGT project focuses on nephroblastoma and breast cancer, basing its works on a master ontology for cancer that is intended to support the automated creation of clinical report forms in order to support clinical trials in cancer genomics research. The @neurIST project focuses on the acquisition and the estimation of the risk for intracranial aneurysms and subarachnoid hemorrhage based on multimodal data, such as cerebral blood flow models or population-based data.
In both environments, it is a challenge to design ontologies capable of addressing the respective broad scopes and of supporting the integration of all available data: In this context, the goal of semantic interoperability seeks to bring about a situation in which all data collected for each individual patient, for each experiment, and for each literature abstract considered relevant for the pertinent domains, has to point to specific nodes (i.e. entity types) in shared domain ontologies. Therefore, the ontologies have to integrate all the various description levels present in the available data (e.g., literature, clinical databases, imaging databases and terminologies).
4 Interfaces of Ontologies
Lessons learned from the ontologies of ACGT [Brochhausen et al. 08] and @neurIST [Boeker et al. 07] have shown that the shortcomings and boundary problems described in section 2 can be alleviated by clearly defining the interfaces between the ontologies and other artifacts that together make up an environment for semantic interoperation. In the following we present a list of six different types of interfaces that an ontology must provide or able to deal with in order for an ontology to be readily useable in real-world scenarios such as the one presented in the last paragraph. We believe that those interfacing techniques – together with the above proposed principles, i.e. the clear separation of concerns of ontology and terminology – can aid in overcoming the problems exposed in this article.
4.1 Clinical Ontologies and Terminologies
Ontologies – in the strict sense of formal ontologies (see section 2.2) – are domain descriptions that are independent of human language, so they need not incorporate any lexical or term information at all. They are theories, like physics. The fact that – for practical reasons – they commonly employ human-readable labels is not a contradiction to this claim. These labels may, but do not have to, coincide with actual domain terms. Usually they do so because ontologies need to be maintained by humans, and are often used by humans in expressing their results without any intervention of a machine, and hence it is obvious that clear natural language labels are needed to allow for the correct use of the ontology nodes they are attached to.
Both the labels and their descriptions for the classes in ontologies should be as precise, unambiguous and self-explanatory as possible. This is often not the case with typically used domain terms.
The interface between domain ontologies on the one hand and domain term lists on the other hand is characterized by a many-to-many relationship: Several terms may be connected to one ontology class due to the phenomena of synonymy and cross-language translation, and polysemy has to be accounted for to link an ambiguous term to more than one ontology class.
For instance, the natural language terms “mamma carcinoma”, “breast cancer”, “Brustkrebs”, and “malignant growth of the female mammary gland” are linked to the same ACGT class in the ontology since they have the same underlying meaning. In contrast to this example, the term “ulcer” points to two distinct nodes in the ontology structure because this term can either refer to the process of ulceration on the one hand or to the pathological structure, the actual result of this process.
4.2 Clinical Ontologies and Upper Ontologies
In our application scenario, ontologies are rooted in upper ontologies. According to [SUO-WG 03] upper ontologies provide generic categories or types suited to address a broad range of domain areas at a high level in a way which can support integration of the underlying data. These upper-level types provide a highly general, clearly formalized structure which can help to consistently represent the entity types in the associated domains. This is further aided by the documentation given alongside the actual upper ontology which documents and explains the respective design principles and further advises ontology developers on how to properly follow those principles in their own ontology development.
It is assumed that an upper ontology can improve (and speed up) both the development process as well as the quality of the actual domain ontologies by providing some consistent and sound top-level framework. Whereas the ACGT ontology uses the Basic Formal Ontology (BFO) [Grenon et al. 04] as its upper level, the @neurIST ontology employs the Descriptive Ontology for Linguistic and Cognitive Engineering (DOLCE) [Gangemi et al. 02]. For an in-depth comparison of these two upper level ontologies (and others) see [Mascardi et al. 07] (see Fig. 2).
Figure 2.
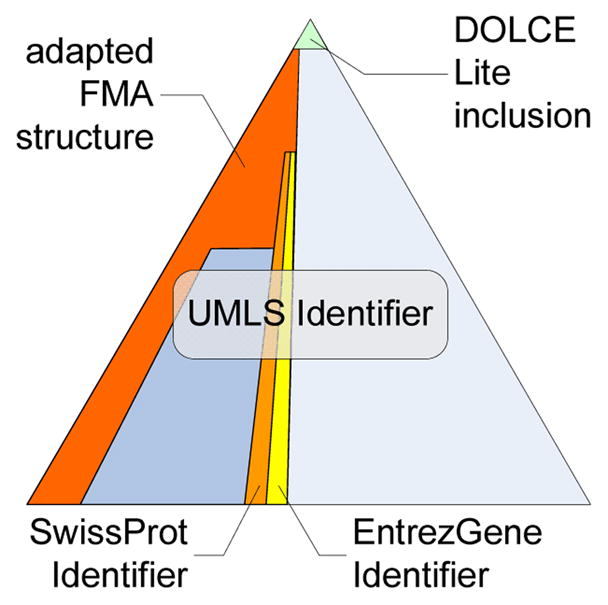
The @neurIST ontology includes DOLCE as upper ontology and adopts the FMA as anatomical reference ontology. SwissProt and EntrezGene ontologies as well as the UMLS terminology is linked via unambiguous identifiers.
4.3 Clinical Ontologies and Non-Ontological Knowledge
As we said before, the domain representations generated in large research projects must extend what can be expressed by ontologies. This extension is referred to as ‘non-ontological knowledge’ in here. Typical examples constitute assertions such as “A treats B” and “C is a risk for D”. Although this knowledge addresses classes at the level of terminology rather than data on individual instances, it is not suitable for inclusion in an ontology since it does not express what holds to be universally true for the given types. In principle, there exist two different ways to represent such kind of knowledge:
The first one follows the classical thesaurus scheme, as employed by the UMLS Metathesaurus, the basic building blocks of which are ‘concept – relation – concept’ triplets. This representation is rather informal and can because of that not be used for logic-based reasoning about classes. Nevertheless the available knowledge can be used for informal reasoning about concepts, e.g. searching for related concepts by graph transversal algorithms or by computing probabilistic associations acquired from training data.
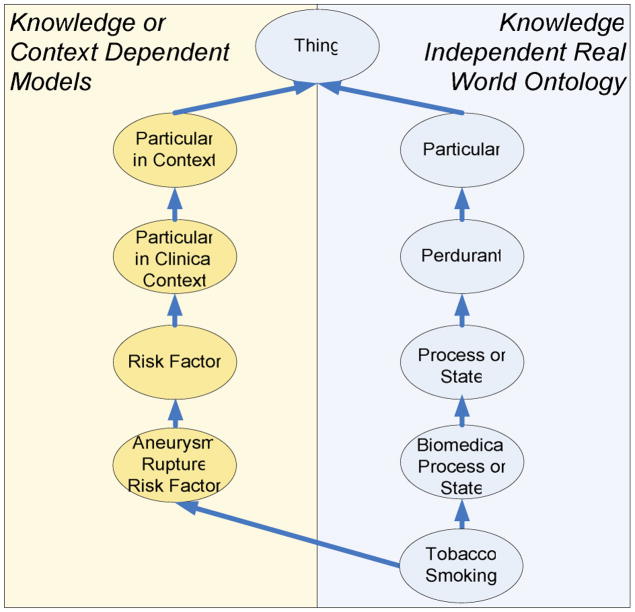
The second solution which has been applied in the context of the @neurIST ontology, consists in a parallel system of non-ontologically grounded, reified classes such as “Suspected Risk Factor for Aneurism Rupture” thus inserting epistemological criteria into the taxonomy. Such categories are generally irrelevant for the correct ontological description of entities but are needed for specific retrieval requirements of ontology users. However, this difference does not affect the fundamental representation formalism. So “Hypertension” a subclass of the above-mentioned class just as it is a subtype of “Cardiovascular Disorder” (see Fig. 3).
Figure 3.
The @neurIST ontology itself is free of any knowledge or context dependent types. The requirement to include classes which represent knowledge dependent types is met by a second hierarchy (Particular in Context) which child classes are reclassified.
4.4 Clinical Ontologies, Information Models and the World
In contradistinction to ontologies, clinical information models can be seen as templates used in Electronic Health Records for the acquisition of clinical data to enable the semantic interoperability within the scope of the specific given clinical information model but not between two or more different clinical information models.
Ideally, clinical information models should be built in such a way as to involve reference to ontologies. Clinical information models offer templates for statements in a medical record. They can encode different things: On the one hand, contextual assertions about concrete ontology instances can be expressed. On the other hand, the existence of such instances can be confirmed, hypothetically assumed with different degrees of certainty, or ruled out. They must be able to cover the whole range of expression like “fever”, “suspected fever”, “history of fever”, or “no fever”. This fact makes clear why clinical information models are necessary and the simple instantiation of ontology classes is not sufficient in clinical documentation. Ontologies provide the types for the particular instances to be recorded in a clinical information model.
This kind of relation has recently received some increased attention in the context of the openEHR Archetypes [Beale et al. 01, Kalra et al. 05], HL7 Version 3 Clinical Document Architecture, and SNOMED CT [Rector et al. 06], and has been further discussed by including experiences from large scale implementation attempts such as the UK Connecting for Health project [HL-7 09].
4.5 Clinical Ontologies and the Ontology Engineer
The actual interface between clinical ontologies and their respective developers is an editing environment that ideally offers support for ontology development and maintenance by a graphical user interface. In this way, the ontology engineer is released from the need to access and edit the ontology’s actual source code, e.g., in the form of the Web Ontology Language (OWL) [OWL 04]).
Most editors allow the user to further describe ontology nodes both with additional textual information and logical definitions. The latter can be used by terminological reasoners to enable automated checking of the structural and (to some extent) semantic correctness of the ontology.
The most prominent currently available ontology editor is Protégé [Protégé 09] which is freely distributed as open-source software and provides (among other things) graphical tree views of the classes and properties with ontologies and special editing facilities for logical constraints on entity types. This editor allows also for the integration external reasoner applications for automatic consistency-checking and classification of the edited ontologies. One of the best-known reasoner applications is Pellet [Sirin et al. 07] which is also available on an open-source basis. Both the ACGT and @neurIST projects employ Protégé in conjunction with Pellet.
A caveat worth mentioning here is that even though those editing tools support the engineer in developing ontologies, they still require the engineer to have quite some proficiency in the underlying technical details and capabilities of the ontology implementation language, such as OWL. Another caveat addresses the maturity of the currently available ontology editing and reasoning tools which have both grown out of academic projects and have not yet reached industry-standard. This has considerable impact on the building and management of large ontologies as well as on the stability and trustworthiness of automated reasoning.
4.6 Clinical Ontologies and the Application Builder
Application builders need a way to programmatically access the content and structure of an ontology to be able to create software systems referring to it, as in the case of ACGT and @neurIST. To reach this goal, more generic application programming interfaces (API) have been designed that abstract away from the actual inner logical structure of the ontologies and present their content via object-oriented (programming language) classes and packages in a way familiar to software developers. Such APIs can subsequently be used by application builders, for example, to programmatically take entity types from the ontology and connect them with external multilingual terminology systems.
Another example is the development of easy-to-use retrieval interfaces for ontological content. It has turned out that current ontology editors such as Protégé, are much too complex for the tasks application builders usually have to perform: Even though they present graphical user interfaces, those interfaces do not make the underlying ontology formalisms transparent enough for application builders who are not proficient in those formalisms. Therefore the ontological content has to be preprocessed and presented in a familiar fashion they can easily relate to. As an example, OWLDoc [Horridge 07] is an open software tool which generates documentation from a given ontology implemented in OWL into HTML format and based on the documentation style familiar to Java programmers.
In the above, we have intentionally avoided the description of any interface between clinical ontologies and the subject of the medical record, i.e., the patient. Furthermore, we have not deliberated on any direct link between the author or the medical record – i.e., the physician – and clinical ontologies. The reason is that such interfaces do not exist in our approach: The relation between the patient and the ontology is an indirect one in that real or hypothetic facts about all aspects of the patient are encoded in the medical record by the physician. The latter is aided by a clinical information model to collect the right data. And finally, the clinical information model links documentation entities to the ontology. Therefore, in the ideal case, the ontology will remain hidden and so transparent to the actual end-user and only provides intelligent support for clinical data acquisition and analysis.
5 Conclusion
Ontologies have become one of the most prominent and important resources in current biomedical informatics research. They are employed for large and multicentric clinical research projects because of their inherent promise to foster semantic interoperability. Ontologies promise to offer both a stable as well as language-independent vocabulary that can help in standardizing and explaining the actual meaning of domain terms.
However, the term ‘ontology’ is often mixed up with related expressions such as ‘terminology’ and ‘thesaurus’, and also with representations of contingent or probabilistic domain knowledge. Database-centered clinical information models that are employed for the recording of patient-specific instance data are, occasionally, also (wrongly) denoted as ontologies.
Aware of the difficulties of creating consensus about the concept of ontologies and aiming at reducing the overlap with other knowledge representation artifacts, we have here introduced a rather restricted notion of clinical ontologies, one that is clearly delimited from the concept of terminology on the one hand, and clinical information model on the other hand. The main distinctive criterion between clinical ontologies and terminologies is in our view the representation of domain entities by the former, against the representation of linguistic entities by the latter.
Clinical ontologies have to be distinguished from clinical information models insofar as the former are restricted to describing and classifying types of domain entities in terms of their generic properties, while the latter provide a description framework for concrete instances of the electronic health record. Such record instances are only indirectly related to entities of the domain itself. Therefore we also propose the introduction of unambiguously defined interfaces between ontologies and other supporting artifacts as well as to the actual application builders to ease the clarification of this distinction.
Acknowledgments
We would like to thank our colleagues and ontology co-developers Susanne Hanser (@neurIST), Mathias Brochhausen (ACGT) and Cristian Cocos (ACGT) for the many helpful discussions surrounding the various topics covered in this article.
The work of Stenzhorn, Schulz, and Boeker on this publication has been carried out jointly within the @neurIST and the ACGT integrated projects funded by the European Commission (IST-027703 and IST-026996).
The work of Smith is supported by the National Institute of Health’s Roadmap for Medical Research, Grant 1 U 54 HG004028.
Footnotes
In the context of this article we do not require the existence of inter-concept relations as a necessary criterion for terminologies.
Contributor Information
Holger Stenzhorn, Email: holger.stenzhorn@uniklinik-freiburg.de, Institute for Medical Biometry and Medical Informatics, University Medical Center Freiburg, Freiburg, Germany and, Institute for Formal Ontology and Medical Information Science, Saarbrücken, Germany.
Stefan Schulz, Email: stschulz@uni-freiburg.de, Institute for Medical Biometry and Medical Informatics, University Medical Center Freiburg, Freiburg, Germany.
Martin Boeker, Email: martin.boeker@uniklinik-freiburg.de, Institute for Medical Biometry and Medical Informatics, University Medical Center Freiburg, Freiburg, Germany.
Barry Smith, Email: phismith@buffalo.edu, Department of Philosophy and Center of Excellence in Bioinformatics and Life Sciences, University at Buffalo, New York, USA and Institute for Formal Ontology and Medical Information Science, Saarbrücken, Germany.
References
- Advancing Clinico-Genomic Trials on Cancer: project website. 2009 March; http://www.eu-acgt.org.
- Integrated Biomedical Informatics for the Management of Cerebral Aneurysms (@neurIST): project website. 2009 March; http://www.aneurist.org.
- Baader F, Calvanese D, McGuinness D, Nardi D, Patel-Schneider P. The Description Logic Handbook: Theory, Implementation and Applications. Cambridge University Press; Cambridge, United Kingdom: 2003. [Google Scholar]
- Boeker M, Stenzhorn H, Kumpf K, Bijlenga P, Schulz S, Hanser S. The @neurIST Ontology of Intracranial Aneurysms: Providing Terminological Services for an Integrated IT Infrastructure. Proc. of AMIA 2007; Chicago, USA. 2007. [PMC free article] [PubMed] [Google Scholar]
- Brochhausen M, Weiler G, Cocos C, Stenzhorn H, Graf N, Doerr M, Tsiknakis M. The ACGT Master Ontology on Cancer - a New Terminology Source for Oncological Practice. Proc. of IEEE CBMS 2008; Jyväskylä, Finland. 2008. [Google Scholar]
- Beale T, Goodchild A, Heard S. EHR Design Principles. London, United Kingdom: 2001. [Google Scholar]
- Bodenreider O, Smith B, Burgun A. The Ontology-Epistemology Divide: A Case Study in Medical Terminology. Proc. FOIS-2006; Torino, Italy. 2004. [PMC free article] [PubMed] [Google Scholar]
- Dolin R, Alschuler L, Boyer S, Beebe C, Behlen F, Biron P, Shabo Shvo A. HL7 Clinical Document Architecture, Release 2. J Am Med Inform Assoc. 2006;13(1):30–9. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi A, Guarino N, Masolo C, Oltramari A, Schneider L. Sweetening Ontologies with DOLCE. Proc. EKAW-2002; Siguenza, Spain. 2002. [Google Scholar]
- Garde S, Knaup P, Hovenga E, Herd S. Towards Semantic Interoperability for Electronic Health Records. Method Inf Med. 2007;46(3):332–343. doi: 10.1160/ME5001. [DOI] [PubMed] [Google Scholar]
- Grenon P, Smith B, Goldberg L. Biodynamic Ontology: Applying BFO in the Biomedical Domain. In: Pisanelli D, editor. Ontologies in Medicine. IOS Press; Amsterdam, Netherlands: 2004. [PubMed] [Google Scholar]
- HL7 Terminfo. 2009 March; http://www.hl7.org/Special/Committees/terminfo.
- Horridge M. OWLDoc - JavaDoc style HTML Documentation for OWL Ontologies. 2007 June; http://www.co-ode.org/downloads/owldoc.
- Ingenerf J, Reiner J, Seik B. Standardized Terminological Services Enabling Semantic Interoperability between Distributed and Heterogeneous Systems. Int J Med Inform. 2001;64(2–3):223–40. doi: 10.1016/s1386-5056(01)00211-8. [DOI] [PubMed] [Google Scholar]
- International Organization for Standardization: ISO 1087-1: Terminology work – Vocabulary – Part 1: Theory and applications. Geneva, Switzerland: 2000. [Google Scholar]
- Jansen L. On Ascribing Dispositions. In: Gnassounou Bruno, Kistler Max., editors. Dispositions and Causal Powers. Ashgate: Aldershot; 2007. pp. 161–177. [Google Scholar]
- Kalra D, Beale T, Heard S. The openEHR Foundation. Stud Health Technol Inform. 2005;115:153–73. [PubMed] [Google Scholar]
- Kayaalp M. Why Do We Need Probabilistic Approaches to Ontologies and the Associated Data?. Proc. Of AMIA Ann Symp; 2005. p. 1005. [PMC free article] [PubMed] [Google Scholar]
- Mascardi V, Cordì V, Rosso P. A Comparison of Upper Ontologies. Genova, Italy: 2007. Technical Report DISI-TR-06-2. [Google Scholar]
- Merriam-Webster, Incorporated: Merriam-Webster’s Medical Dictionaryu. 2008 March; http://medical.merriam-webster.com.
- Open Biological Ontologies (OBO) Foundry. 2008 March; http://obofoundry.org.
- W3C, Web Ontology Language (OWL) Overview. 2004 February; http://www.w3.org/TR/owl-features.
- Ontology Protégé., editor. 2009 March; http://protege.stanford.edu.
- On What There Is. Review of Metaphysics. (1948)
- Rector A, Zanstra P, Solomon W, Rogers J, Baud R. Reconciling Users Needs and Formal Requirements: Issues in Developing Re-Usable Ontology for Medicine. IEEE Transactions on Information Technol in BioMedicine. 1999;2(4):229–242. doi: 10.1109/4233.737578. [DOI] [PubMed] [Google Scholar]
- Rector A, Qamar R, Marley T. Binding Ontologies and Coding Systems to Electronic Health Records and Messages. Proc. KR-MED-2006; Baltimore, USA. 2006. [Google Scholar]
- Rosse C, Mejino JA. Reference Ontology for Biomedical Informatics: The Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Rossi-Mori A, Consorti F. Exploiting the Terminological Approach from CEN/TC251 and GALEN to Support Semantic Interoperability of Healthcare Record Systems. Int J Med Inform. 1998;48(1–3):111–124. doi: 10.1016/s1386-5056(97)00116-0. [DOI] [PubMed] [Google Scholar]
- Sirin E, Parsia B, Cuenca Grau B, Kalyanpur A, Katz Y. Pellet: A Practical OWL-DL Reasoner. Journal of Web Semantics. 2007;5:2. [Google Scholar]
- Smith B. Against Fantology. In: Reicher M, Marek J, editors. Experience and Analysis. 2005. pp. 153–170. [Google Scholar]
- Smith B, Kusnierczyk W, Schober D, Ceusters W. Towards a Reference Terminology for Ontology Research and Development in the Biomedical Domain. Proc. KR-MED-2006; Baltimore, USA. 2006. [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard C, Bug W, Ceusters W, Goldberg L, Eilbeck K, Ireland A, Mungall C, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S, Scheuermann R, Shah N, Whetzel P, Lewis S The OBI Consortium. The OBO Foundry: Coordinated Evolution of Ontologies to Support Biomedical Data Integration. Nature Biotechnology. 2007;25:1251 – 1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Health Terminology Standards Development Organisation. SNOMED-CT. 2009 March; http://www.ihtsdo.org/snomed-ct/
- Standard Upper Ontology Working Group (SUO-WG) 2003 http://suo.ieee.org.
- National Library of Medicine, Unified Medical Language System (UMLS) Bethesda, MD, USA: 2009. http://www.nlm.nih.gov/research/umls. [Google Scholar]
- Uschold M, King M. Ontologies: Principles, Methods, and Applications. Knowledge Eng Rev. 1996;11(2):93–155. [Google Scholar]
- World Health Organization, International Statistical Classification of Diseases (ICD) Geneva, Switzerland: 2009. http://www.who.int/classifications/icd. [Google Scholar]