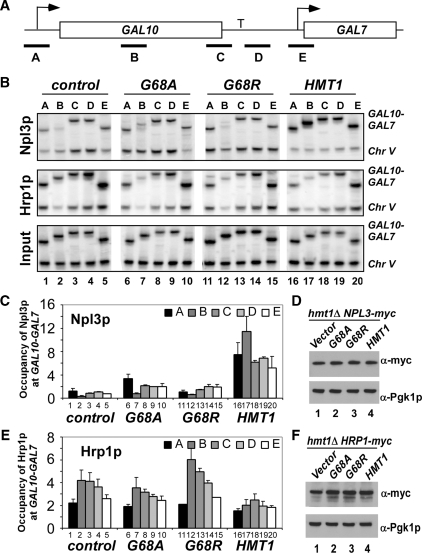
Figure 3.
Loss of HMT1 affects Npl3p and Hrp1p occupancies at activated GAL10-GAL7. (A) Diagram of the GAL10-GAL7 locus, with PCR primer pairs indicated with black bars and terminator labeled as ‘T’. (B) ChIP analysis of Npl3p (upper panel) and Hrp1p (middle panel) occupancies across the GAL10-GAL7 locus. ChIP analysis of Myc-Npl3p or Myc-Hrp1p occupancies was conducted using antimyc antibodies as described in Figure 2, except using primers to amplify different regions of GAL10-GAL7 in panel A and to amplify non-transcribed region of Chr V was conducted to control for non-specific immunoprecipitation. Upper panel, transformants of hmt1Δ NPL3-myc strain (CMY201) containing empty vector (Lanes 1–5), or plasmids harboring hmt1-G68A (Lanes 6–10), hmt1-G68R (Lanes 11–15) or HMT1 (Lanes 16–20) were grown in SC-LEU galactose medium. Middle panel, transformants of hmt1Δ HRP1-myc strain (CMY203) harboring the same plasmids described for the upper panel were analyzed as described earlier. Lower panel: Analysis of input chromatin samples for the CMY201 transformants. (C) Quantification of Npl3p occupancy measured at the indicated positions. The ratios of experimental-to-control signals in the immunoprecipitate samples (B, upper panel) were normalized for the corresponding ratios for input samples (B, lower panel) to yield the occupancy values. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (D) Possible effect of mutation or deletion of HMT1 on expression of Myc-tagged Npl3p was analyzed by western blot analysis of WCEs of the relevant strains with antiMyc and antiPgk1p antibodies (to provide a loading control). (E) Quantification of Hrp1p occupancy measured at the indicated positions. The ratios of experimental to-control signals in the immunoprecipitate samples (B, middle panel) were normalized for the corresponding ratios for input samples (B, lower panel) to yield the occupancy values. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (F) Possible effect of mutation or deletion of HMT1 on expression of Myc-tagged Hrp1p was analyzed by western blot analysis of WCEs of the relevant strains with antiMyc and antiPgk1p antibodies (to provide a loading control).