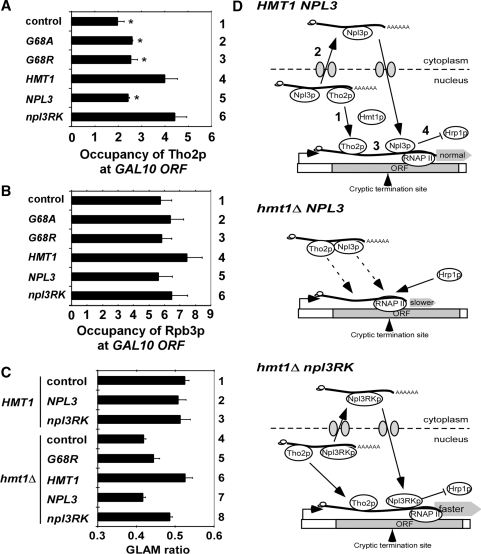
Figure 5.
Tho2p recruitment and gene length-dependent transcription defects in hmt1Δ cells are suppressed by npl3RK, and a hypothetical model for the role of Hmt1p in recycling Tho2p and Npl3p from mRNPs to sites of transcription. ChIP analysis of Tho2p occupancy at the 5′ region of GAL10 gene (region B in Fig. 3A) in hmt1Δ THO2-myc13 (CMY202) cells. Cells transformed with empty vector, or plasmids containing hmt1-G68A, hmt1-G68R, HMT1, NPL3 or npl3RK were grown in SC-LEU with 2% galactose and ChIP analysis was conducted using antiMyc antibodies (to measure Tho2p occupancy) or antibodies against Rpb3p (to measure RNAP II occupancy). Non-coding region on chromosome V (Chr V) was amplified as control for non-specific immunoprecipitation. The occupancies were quantified and plotted for Tho2p (A) and Rpb3p (B). Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. The occupancies of Tho2p at the GAL10 ORF are significantly lower in control (hmt1Δ), G68A and G68R versus HMT1 cells, and in cells containing extra copies of NPL3 versus extra copies of npl3RK, as judged by the Student’s t-test, with P < 0.01(*). (C) HMT1 (BY4741) and hmt1Δ (3171) cells transformed with reporter plasmid pSCH209-LAC4 plus plasmids harboring hmt1-G68A, NPL3, npl3RK or empty vector were grown in SC-LEU-URA with 2% galactose. HMT1 (BY4741) and hmt1Δ (3171) cells harboring pSCH202 were analyzed as controls. Acid phosphatase activity was measured in cell lysates and the GLAM ratio was calculated as the ratio of activities in transformants harboring pSCH209-LAC4 versus those containing pSCH202. Results are the averages from three independent experiments and error bars indicate SD (D) Model depicting how Hmt1p regulates transcription elongation and termination by methylating Npl3p. Upper panel: In WT cells, methylation of Npl3p by Hmt1p allow dissociation the TREX(Tho2p)−Npl3p−mRNP to facilitate (1) TREX(Tho2p) recruitment to sites of transcription and (2) nuclear export of the Npl3p−mRNP. Subsequent nuclear import and methylation of Npl3p by Hmt1p enables its recruitment to sites of transcription (3). Npl3p and TREX both stimulate elongation and, by impeding recruitment of Hrp1p (4), Npl3p prevents transcription termination at cryptic terminators. Middle panel, in hmt1Δ NPL3 cells, the failure to methylate Npl3p causes Tho2p (TREX) and Npl3p to be sequestered in mRNPs that are not exported efficiently; the ensuing impaired recruitment of TREX(Tho2p) and Npl3p to sites of transcription reduces the rate of elongation, and the resulting increased Hrp1p occupancy enables transcription termination at cryptic terminators. Lower panel, in hmt1Δ npl3RK cells, TREX(Tho2p)-Npl3RKp-mRNP can dissociate independently of Npl3p methylation to recycle TREX(Tho2p) and Npl3RKp back to sites of transcription where they stimulate elongation and block termination at cryptic terminators, as occurs in WT cells.