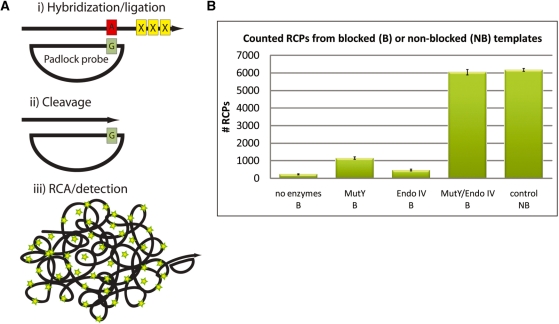
Figure 4.
Removal of blocked 3′-end with MutY/Endo IV for RCA. (A) The mechanism of removal of the 3′ blocking end with MutY/Endo IV. (i) An oligonucleotide with a blocked 3′-end serves as a target for padlock hybridization and ligation. The duplex formed between the probe/target creates a G:A mismatch. (ii) Recognition and cleavage of mismatch by MutY glycosylase/Endo IV resulting in removal of the blocked 3′-end. (iii) RCA and detection of RCPs. (B) Results from quantification of RCPs with the digital RCA technique. Briefly the procedure involves hybridization of fluorescently labeled detection oligonucleotides to the RCPs followed by pumping the solution through a microfluidic channel that is mounted on a fluorescent microscope. The imaged RCPs show up as distinct fluorescent spots that are quantified using imaging software. The resulting number of RCPs was counted in the different blocked (B) or non-blocked (NB) samples: negative control (no MutY/Endo IV added) (B), MutY (B), Endo IV (B), MutY/Endo IV (B) and positive control (NB). Error bars indicate standard deviations.