Table 1.
Parameters used in the analysis
| Parameter | Unit | Description | Relation to other parameters |
|---|---|---|---|
| L | (nt) | Track length (nt = nucleotide unit) | |
| b | (nt) | Binding site size | |
| C | (nt−1) | Mechanochemical coupling ratio ([NTP hydrolyzed]/ [enzyme]/nt traveled) | C = ka/ktrans = c/m |
| ka | (s−1) | Steady-state NTP hydrolysis rate during translocation ([NTP hydrolyzed]/ [enzyme]/s) | ka = Cktrans = c(kt + kd); (kt + kd = net rate constant of kinetic step) |
| ktrans | (nt s−1) | Macroscopic rate of translocation | ktrans = ka/C = m(kt + kd) |
| c | – | Coupling stoichiometry ([NTP hydrolyzed]/ [enzyme]/kinetic step) | c = mka/ktrans = Cm |
| m | (nt) | Kinetic step size (nt traveled/kinetic step) | m = cktrans/ka = c/C |
| P | – | Kinetic processivity (probability of taking the next kinetic step) | P = kt/(kt + kd) |
| Pmacr | – | Macroscopic processivity, suitable for calculation of: Mean number of NTP molecules consumed in a single run (at infinite track length) = Pmacr/(1–Pmacr) Mean number of nucleotides travelled in a single run (at infinite track length) = Pmacr/(1–Pmacr)/C |
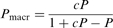 [see also Equation (10)] [see also Equation (10)] |
