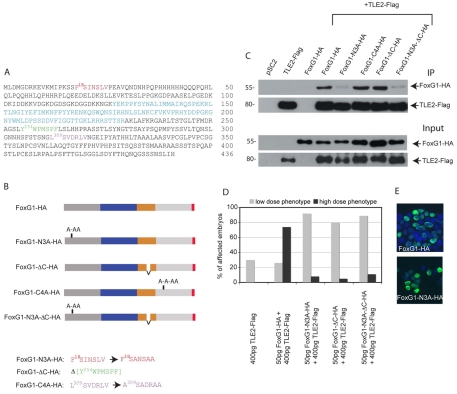
Fig. 6.
Motifs in FoxG1 that are responsible for the binding and functional cooperativity of FoxG1 and TLE2. (A) Amino acid sequence of X. laevis FoxG1 and potential TLE binding motifs. The DNA-binding domain is shown in blue, the N-terminal eh1 motif is highlighted in red, the C-terminal WRPW-like motif in green and the cryptic eh1 motif in purple. (B) Schematic representation of the FoxG1 mutants. The N-terminal domain is shown in dark grey, the DNA-binding domain in blue, the potential TLE1 interaction domain in orange and the HA epitope in red. Beneath is shown the sequence of mutated motifs (same colour coding as in A). (C) Immunoprecipitation (IP) of HEK 293T cells transfected with FoxG1-HA or FoxG1 mutants together with TLE2-Flag, showing that mutation of the N-terminal eh1 motif (FoxG1-N3A-HA), either alone or simultaneously with the C-terminal motif (FoxG1-N3A-ΔC-HA), leads to loss of binding to TLE2. The top two panels show IP with anti-Flag followed by western blotting with anti-HA and anti-Flag. Lower panels show the input (pre-IP lysates) analysed by western blotting with anti-Flag and anti-HA. (D) The phenotypes of embryos injected with the indicated amounts of X. tropicalis TLE2 and X. laevis FoxG1 or FoxG1 mutant mRNA were scored as low-dose or high-dose as in Fig. 5. TLE2 does not cooperate with any of the three mutants to convert a low-dose FoxG1 injection into a high-dose phenotype. (E) HEK 293T cells transfected with either FoxG1-HA or FoxG1-N3A-HA. Both proteins are nuclear.