Figure 3. Shed vesicles exhibit oncosome activity.
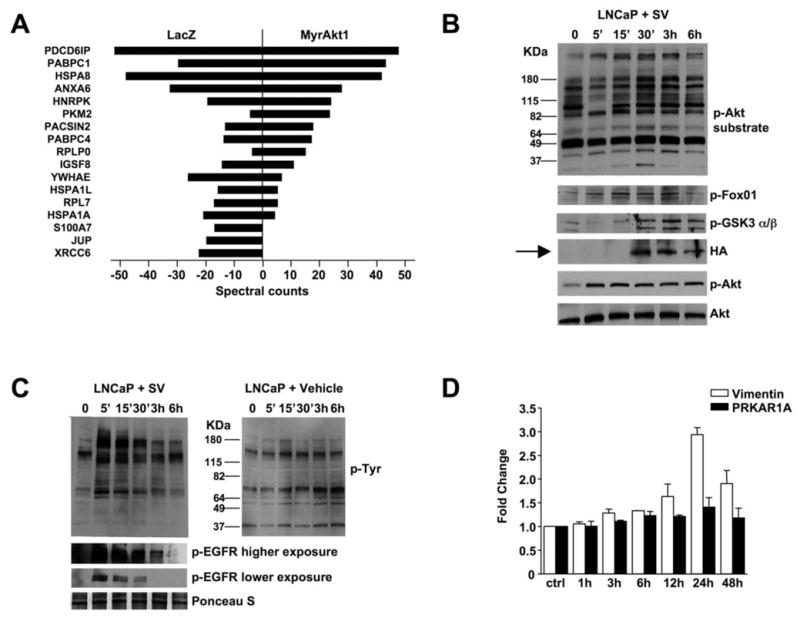
(A) Proteins identified by tandem mass spectrometry in bleb material and quantified using spectral counting. PDCD6IP (Programmed Cell Death 6 Interacting Protein), PABPC1 (Poly(A) Binding Protein, Cytoplasmic 1), HSPA8 (Heat Shock 70kDa Protein 8), ANXA6 (Annexin A 6), hnRNP-K (Heterogeneous Nuclear Ribonucleoprotein K, PKM2 (Pyruvate Kinase M2), PACSIN2 (Protein Kinase C and Casein Kinase Substrate in Neurons 2), PABPC4 (Poly(A) Binding Protein, Cytoplasmic 4), RPLPO (Ribosomal Protein Large P0-like Protein), IGSF8 (Immunoglobulin Superfamily, member 8), YWHAE (Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase Activation Protein, Epsilon Polypeptide), HSPA1L (Heat Shock 70kDa Protein 1-Like), RPL7 (Ribosomal Protein L7), HSPA1A (Heat Shock 70kDa protein 1A), S100A7 (S100 calcium binding protein A7), JUP (Junction Plakoglobin), XRCC6 (X-ray Repair Complementing defective repair in Chinese hamster cells 6). (B) LNCaP/LacZ cells were exposed to SV obtained from EGF-treated LNCaP/MyrAkt1 cells. Blotting of whole cell lysates is shown. (C) LNCaP/LacZ exposed to 20 μg of SV or vehicle from EGF-treated LNCaP/MyrAkt1 cells and assessed for p-Tyr (upper panels). The lower panels show the results of blotting with p-EGFR antibody (p-Tyr1068). (D) WPMY-1 cells were exposed to LNCaP/MyrAkt1 derived SV for the indicated times. Vimentin mRNA was quantified by qRT-PCR in recipient cells, and normalized using GAPDH. An irrelevant gene, PRKAR1A, was used as control.
