Fig. 1.
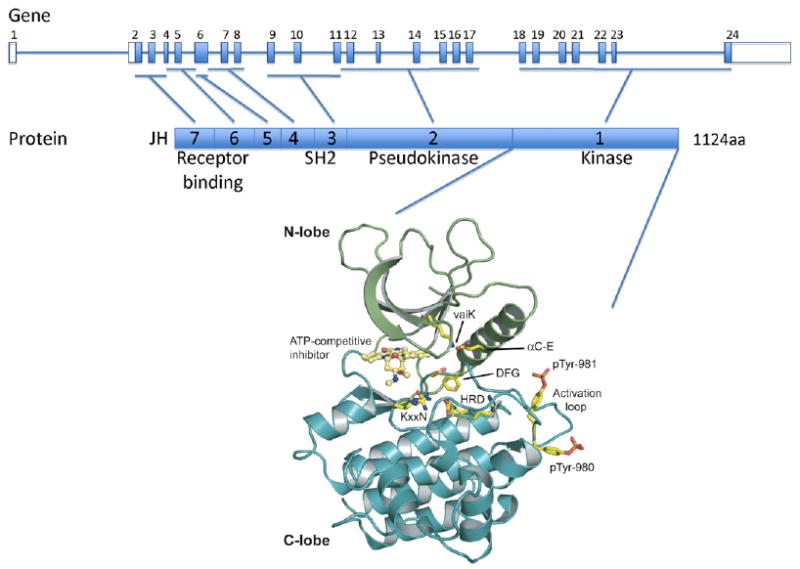
Structure of the JAK3 gene and protein. The human JAK3 genomic structure is shown in the upper panel and the JAK3 protein structure is represented in the middle panel. JAK3 is comprised of 7 JAK homology (JH) domains. JH1 contains the C terminus kinase domain; JH2 contains a pseudokinase domain and the alanine 572 residue which mutation to valine leads to constitutive activation of the kinase in acute megakaryoblastic leukemia and cutaneous T cell lymphoma. The N terminus region (JH6 and JH7) is critical for γc receptor binding and signal transduction. Y100C mutation found in SCID patients abolishes the interaction between JAK3 and the common γc chain. A cartoon representation of the JH1 domain of Jak3 (PDB code: 1YVJ) is shown (N-lobe is in green and C-lobe in blue) in the lower panel. A small molecule staurosporine analogue is seen bound in the kinase catalytic cleft in yellow. Conserved kinase domain residues are shown in stick representation and indicated HRD, KxxN, DFG, αC-E, vaiK and are important for orientation of the substrate residue hydroxyl group (HRD), γ-phosphate neutralization (KxxN), α- and β- phosphate positioning (αC-E, vaiK), metal binding (DFG), and conformational transition between active and inactive states (DFG). Human Jak3 varies from canonical kinase domains with substitutions of the KxxN and vaiK motifs to xxRN and vavK, both commonly seen substitutions. Phosphotyrosines pTyr-980 and pTyr-981 critical for JAK3 activity are shown in stick representation. Figure was made using Pymol (www.pymol.org).
