Figure 1.
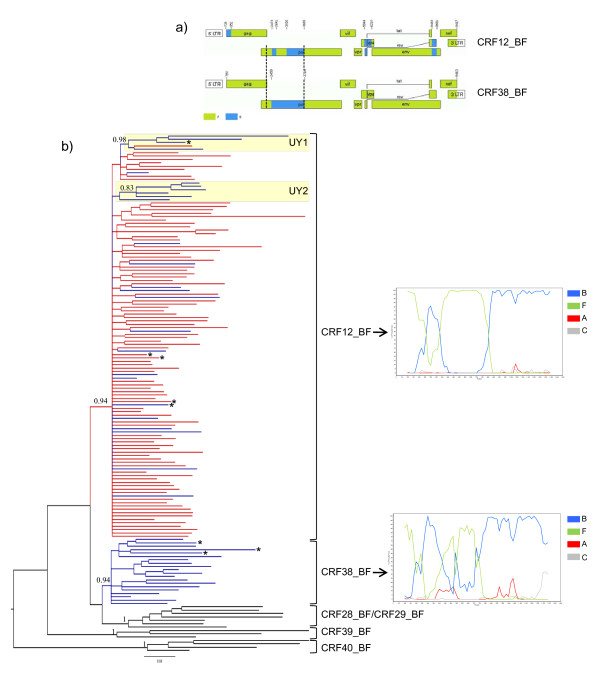
Virus analyses. a) Genomic mosaic structure of CRF12_BF and CRF38_BF viruses. Green, subtype F1; blue subtype B; white, unknown subtype. Numbers above breakpoints refer to nucleotide positions in the HXB2 genome. Vertical dotted lines indicate the pol gene fragment (nucleotides 2266-3705) used in the present study. b) Majority-rule Bayesian consensus tree of the pol gene of HIV-1 CRFs_BF circulating in Argentina (red), Uruguay (blue), and Brazil (black). Posterior probability values are indicated only at key nodes. Brackets indicate the monophyletic clusters formed by each CRF. Boxes indicate the two Uruguayan sub-cluters identified within the CRF12_BF clade. Positions of the full-length characterized CRF12_BF and CRF38_BF reference sequences are marked with asterisks. The tree was rooted on midpoint and horizontal branch lengths are drawn to scale with the bar at the bottom indicating 0.03 nucleotide substitutions per site. Representative bootscanning plots of the pol gene fragment of CRF12_BF (A32879) and CRF38_BF (UY03_3389) reference sequences are depicted on the right. Reference sequences used for these analyses were as follows: subtype B (BZ126, blue), subtype F1 (BZ167, green), subtype C (92BR025, gray) and subtype A1 (U455, red).