Table 3. The 50 genes most distant to the origin of the coordinates space 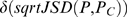 ×
×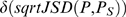 .
.
| Probe Set ID | Gene Symbol | Gene Title | δ(sqrtJSD (P,Pc)) | δ(sqrtJSD (P,Ps)) | Dist O | Ref (ADG) |
| 206481_s_at | LDB2 | LIM domain binding 2 | −0.7988 | 0.7427 | 1.0907 | |
| 219736_at | TRIM36 | tripartite motif-containing 36 | −0.8077 | 0.7242 | 1.0848 | |
| 200650_s_at | LDHA | lactate dehydrogenase A | −0.8210 | 0.6984 | 1.0778 | |
| 205113_at | NEFM | neurofilament, medium polypeptide 150kDa | −0.7448 | 0.7742 | 1.0743 | [379], [649], [672], [673], [674], [675], [676], [677], [678], [679], [680], [681], [682], [683], [684], [685], [686], [687], [688], [689], [690], [691], [692], [693], [694], [695], [696], [697], [698], [699], [700], [701], [702] |
| 202656_s_at | SERTAD2 | SERTA domain containing 2 | 0.7343 | −0.7827 | 1.0732 | |
| 203798_s_at | VSNL1 | visinin-like 1 | −0.7093 | 0.7923 | 1.0634 | [565], [566], [638], [641], [642], [703], [704], [705] |
| 205352_at | SERPINI1 | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | −0.7432 | 0.7496 | 1.0555 | [706], [707], [708], [709], [710], [711], [712], [713], [714] |
| 217367_s_at | ZHX3 | zinc fingers and homeoboxes 3 | 0.7677 | −0.7129 | 1.0477 | |
| 209915_s_at | NRXN1 | neurexin 1 | −0.7282 | 0.7496 | 1.0451 | |
| 221805_at | NEFL | neurofilament, light polypeptide 68kDa | −0.7153 | 0.7552 | 1.0402 | [715], [716], [717] |
| 213366_x_at | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −0.7302 | 0.7327 | 1.0344 | |
| 203340_s_at | SLC25A12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | −0.7141 | 0.7444 | 1.0315 | [718] |
| 213967_at | RALYL | RALY RNA binding protein-like | −0.6786 | 0.7758 | 1.0307 | |
| 215161_at | CAMK1G | calcium/calmodulin-dependent protein kinase IG | −0.7819 | 0.6682 | 1.0285 | |
| 218204_s_at | FYCO1 | FYVE and coiled-coil domain containing 1 | 0.7250 | −0.7222 | 1.0233 | |
| 213222_at | PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) | −0.7694 | 0.6738 | 1.0227 | [719], [720], [721], [722], [723], [724] |
| 200925_at | COX6A1 | cytochrome c oxidase subunit VIa polypeptide 1 | −0.7532 | 0.6883 | 1.0204 | |
| 38037_at | HBEGF | heparin-binding EGF-like growth factor | 0.7222 | −0.7194 | 1.0193 | [725] |
| 209481_at | SNRK | SNF related kinase | −0.7048 | 0.7331 | 1.0169 | |
| 201412_at | LRP10 | low density lipoprotein receptor-related protein 10 | 0.6964 | −0.7399 | 1.0161 | |
| 202941_at | NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | −0.6984 | 0.7379 | 1.0160 | |
| 205383_s_at | ZBTB20 | zinc finger and BTB domain containing 20 | 0.6774 | −0.7569 | 1.0157 | [726], [727] |
| 206384_at | CACNG3 | calcium channel, voltage-dependent, gamma subunit 3 | −0.7778 | 0.6516 | 1.0147 | |
| 218888_s_at | NETO2 | neuropilin (NRP) and tolloid (TLL)-like 2 | −0.7899 | 0.6246 | 1.0070 | |
| 212214_at | OPA1 | optic atrophy 1 (autosomal dominant) | −0.7194 | 0.7024 | 1.0054 | [728], [729], [730], [731], [732], [733], [734], [735], [736], [737], [738], [739], [740], [741], [742] |
| 218456_at | CAPRIN2 | caprin family member 2 | −0.7915 | 0.6186 | 1.0046 | |
| 211307_s_at | FCAR | Fc fragment of IgA, receptor for | 0.7297 | −0.6886 | 1.0033 | |
| 202724_s_at | FOXO1 | forkhead box O1 | 0.6875 | −0.7270 | 1.0006 | [743], [744], [745] |
| 219145_at | LPHN1 | latrophilin 1 | −0.7293 | 0.6826 | 0.9989 | [746] |
| 205711_x_at | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −0.7153 | 0.6968 | 0.9986 | |
| 55662_at | C10orf76 | chromosome 10 open reading frame 76 | 0.7632 | −0.6420 | 0.9973 | |
| 211978_x_at | PPIA | peptidylprolyl isomerase A (cyclophilin A) | −0.7363 | 0.6726 | 0.9972 | [747], [748], [749], [750] |
| 210016_at | MYT1L | myelin transcription factor 1-like /// hypothetical protein LOC100134306 | −0.7577 | 0.6395 | 0.9915 | |
| 204072_s_at | FRY | furry homolog (Drosophila) | −0.7456 | 0.6512 | 0.9899 | |
| 219497_s_at | BCL11A | B-cell CLL/lymphoma 11A (zinc finger protein) | −0.6843 | 0.7117 | 0.9873 | |
| 201125_s_at | ITGB5 | integrin, beta 5 | 0.7323 | −0.6613 | 0.9867 | |
| 211765_x_at | PPIA | peptidylprolyl isomerase A (cyclophilin A) | −0.7230 | 0.6714 | 0.9866 | [747], [748], [749], [750] |
| 214057_at | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 0.7722 | −0.6137 | 0.9864 | [751] |
| 211839_s_at | CSF1 | colony stimulating factor 1 (macrophage) | 0.8120 | −0.5590 | 0.9858 | [752], [753], [754], [755], [756], [757], [758], [759], [760], [761], [762], [763], [764], [765], [766], [767], [768], [769], [770], [771], [772], [773], [774], [775], [776], [777], [778], [779], [780], [781], [782] |
| 205551_at | SV2B | synaptic vesicle glycoprotein 2B | −0.6915 | 0.7008 | 0.9846 | [66] |
| 219167_at | RASL12 | RAS-like, family 12 | 0.6226 | −0.7605 | 0.9828 | |
| 214393_at | RND2 | Rho family GTPase 2 | 0.7051 | −0.6799 | 0.9796 | |
| 212899_at | CDC2L6 | cell division cycle 2-like 6 (CDK8-like) | 0.7319 | −0.6504 | 0.9791 | |
| 220615_s_at | MLSTD1 | male sterility domain containing 1 | −0.6665 | 0.7149 | 0.9774 | |
| 201681_s_at | DLG5 | discs, large homolog 5 (Drosophila) | 0.7093 | −0.6706 | 0.9761 | |
| 208195_at | TTN | titin | 0.7173 | −0.6617 | 0.9759 | [783] |
| 202457_s_at | PPP3CA | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | −0.7661 | 0.6016 | 0.9741 | |
| 214150_x_at | ATP6V0E1 | ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 | 0.6230 | −0.7488 | 0.9741 | |
| 204743_at | TAGLN3 | transgelin 3 | −0.6952 | 0.6802 | 0.9726 | |
| 213197_at | ASTN1 | astrotactin 1 | −0.7069 | 0.6673 | 0.9721 |
The column “Dist O” shows the Euclidean distance from the origin for each gene. If the gene has a known relation with AD (ADG), the reference's codes are display in column “Ref ADG”.
