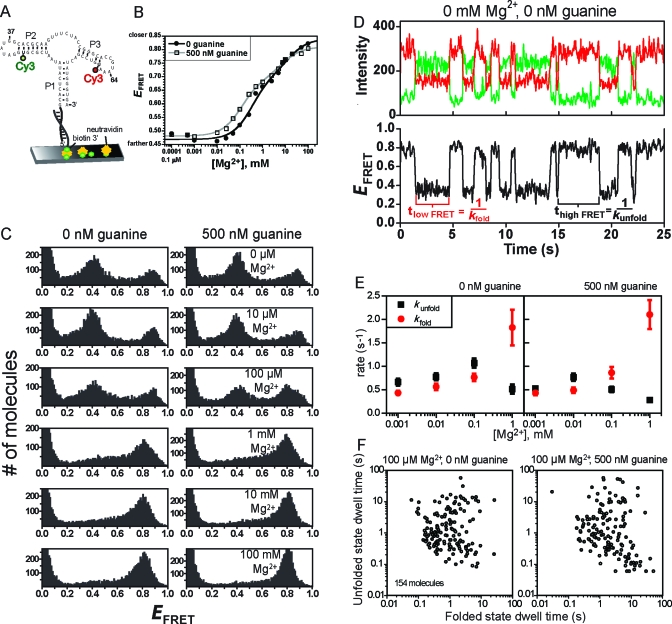
Figure 4.
Ensemble and single-molecule FRET analyses of the P2−P3 aptamer domain variant. (A) Design of P2−P3 for smFRET analysis. For ensemble FRET, the neutravidin surface was omitted. (B) Ensemble FRET titration of P1−P2 with Mg2+ at 0 or 500 nM guanine. Each curve was fitted to a standard two-component titration curve with a Hill coefficient of 1 for each component (39). (C) smFRET histograms for P2−P3. (D) Representative single-molecule smFRET time traces for P2−P3. Time traces at other Mg2+ and guanine concentrations (up to 1 mM Mg2+) were qualitatively similar, although the quantitative dwell time values varied slightly (see panel E). (E) Plots of rate constants kfold and kunfold as determined from single-molecule dwell times. Error bars represent the standard error from averaging rates of all molecules under the indicated buffer condition. (F) The average duration of the high-FRET (folded) and low-FRET (unfolded) dwell times of 154 P2−P3 molecules in 100 μM Mg2+ reveals broad heterogeneity. Inclusion of guanine did not considerably change the heterogeneity. Scatter plots at other Mg2+ concentrations are shown in Figure S5 of the Supporting Information.