Figure 3. Current working model of the genes required for myoblast fusion in Drosophila.
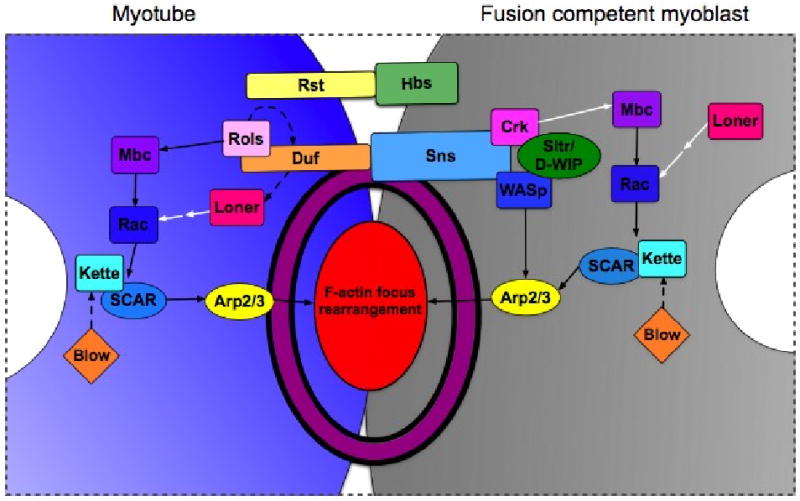
A simplified model that has been updated to reflect recently identified fusion genes and to illustrate the conservation of proteins to the zebrafish and mouse myoblast fusion paradigms. Note that nuclei are in white, myotube in is grey and the FCM is in blue. The actin focus and FuRMAS are depicted as a red oval and a purple ring, respectively. Rectangles represent proteins that have a conserved role in myoblast fusion in multiple systems. Ovals represent proteins with known homologs in vertebrates that have no role in fusion described to date. Diamonds represent proteins that have no known homologs in vertebrates. Solid arrows denote well-characterized biochemical interactions, dashed arrows indicate genetic and/or suggested biochemical interactions and white arrows designate interactions that are suggested from work on orthologous proteins in other contexts. While this depiction suggests specific interacting partners for each transmembrane protein, there is evidence that this may not be the case as interactions between Duf and Hbs have been shown to mediate cell adhesion in vitro. Additionally, the reader is cautioned that, although strong biochemical exists for Mbc-mediated Rac activation and for the Rac→ SCAR complex→ Arp2/3 pathway, there is not yet evidence for a complete pathway linking Mbc to Arp2/3-dependent actin polymerization.
