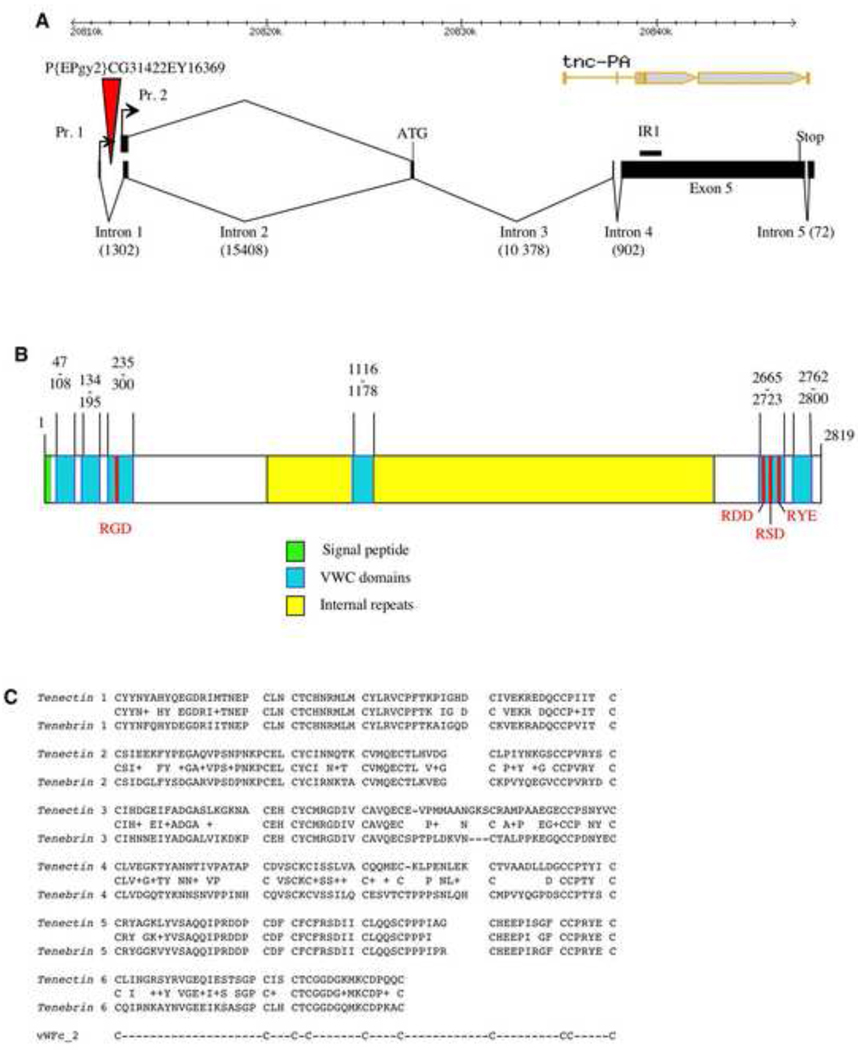
Fig. 1.
tenectin gene organization. (A) Schematic of the tenectin gene showing the start and stop codons and intron splice sites for the two RNAs transcribed from 2 promoters (Pr. 1 and Pr. 2). The two transcripts contain identical translated sequences. The positions of the P{EPgy2}CG31422EY16369 element and sequence used for making the inverted repeat RNAi construct (IR1) are marked. For comparison, the flybase gene structure, with errors in intron/exon positions, is presented at the top of the figure (tnc-PA). (B) Schematic of the deduced protein showing the signal peptide, the RGD motifs, von Willebrand Factor type-c (VWC) domains and internal repeats. (C) Sequence alignments of VWC domains in tenectin and tenebrin. Consensus cysteines are also shown.