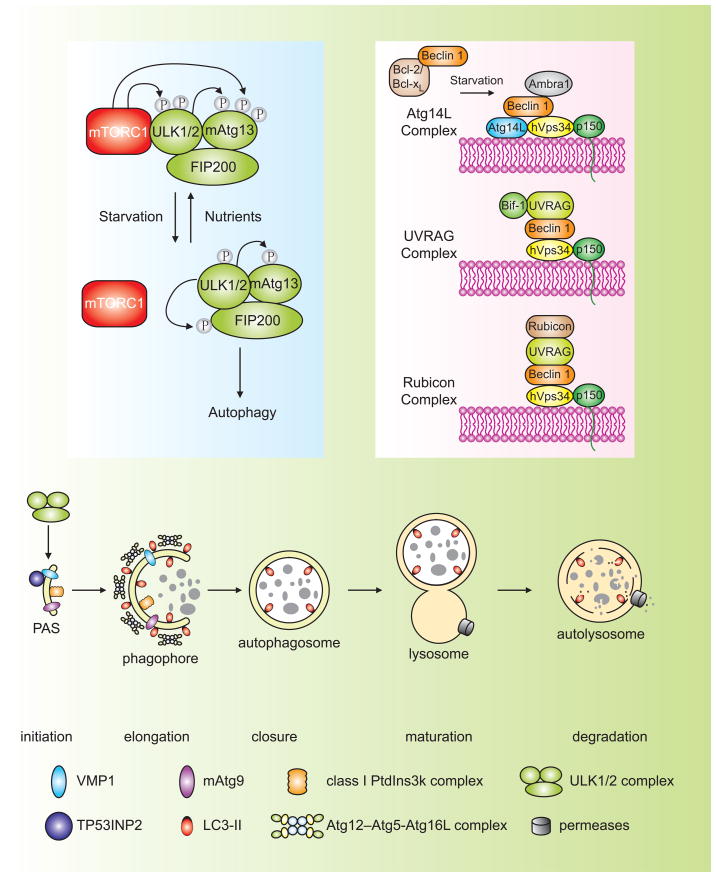
Figure 1. Schematic depiction of the autophagy pathway and its core molecular machinery in mammalian cells.
Mammalian autophagy proceeds through a series of steps, including initiation at the PAS (phagophore assembly site), elongation and expansion of the phagophore, closure and completion of the autophagosome, autophagosome maturation via docking and fusion with an endosome and/or lysosome, breakdown and degradation of the autophagosome inner membrane and cargo, and recycling of the resulting macromolecules. Regulatory components for autophagy induction include the ULK1 and ULK2 complexes that contain various Atg proteins (light blue box at left) that are required for autophagy. The association of mTORC1 with this complex and the activity of mTORC1 depend on the nutrient status. Under nutrient-rich conditions, mTORC1 is associated with the ULK1 and ULK2 complexes, and phosphorylates ULK1, ULK2, and mAtg13; upon inactivation of mTORC1 by nutrient starvation, mTORC1 disassociates, mAtg13, ULK1 and ULK2 are partially dephosphorylated, and activation of ULK1 and ULK2 promotes phosphosphorylation of FIP200. There are at least three class III PtdIns3K complexes (light red box at right), that are involved in autophagosome formation or clearance. The Atg14L (Atg14L-Beclin 1-hVps34-p150) and UVRAG (UVRAG-Beclin 1-hVps34-p150) complexes are required for autophagy, whereas the Rubicon complex (Rubicon-UVRAG-Beclin 1-hVps34-p150) negatively regulates autophagy. Ambra1 and Bif-1 are essential for induction of autophagy, through direct interaction with Beclin 1 and UVRAG, respectively, whereas Bcl-2 binds to Beclin 1 and disrupts the Beclin 1-associated hVps34 complex, thereby inhibiting autophagy.