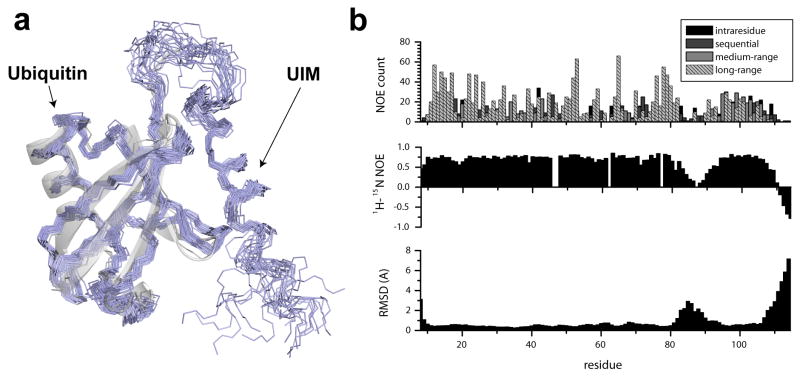
Figure 1. Solution structure of the Ubiquitin/UIM fusion protein.
(a) An overlay of the 20 lowest energy conformations of the converged NMR ensemble (PDB ID 2kdi). A superposition of the x-ray structure of Ubiquitin (PDB ID iubq) is shown to illustrate perturbations in the structure induced by UIM binding. (b) Residue-specific structural statistics for the UIM/Ubiquitin complex. NOE constraint densities per residue (top) are compared to the 1H-induced 15N NOE (middle) and residue-specific RMSD values for the NMR ensemble. Under restrained regions in the ensemble (bottom) coincide with the regions of increased plasticity (middle) as illustrated by reduced 1H-induced 15N NOE and NOE density. Residue numbering includes an initial 9 residue-long Histidine-containing sequence.