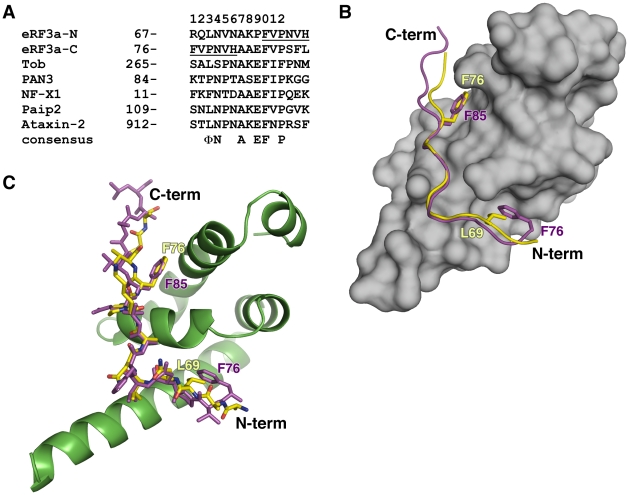
Figure 1. Overlapping PAM2 motifs of eRF3.
(A) Sequence of MLLE-binding PAM2 motifs. PAM2-N and PAM2-C of eRF3 are aligned against sequences from Tob (transducer of Erb1), poly(A) specific ribonuclease 3 (PAN3), PABP-interacting protein 2 (Paip2) and Ataxin-2. Residues in the overlap are underlined. The consensus of most conserved residues that contribute to the MLLE/PAM2 binding is shown: Φ represents a hydrophobic residue [12]. (B) Overlaid structures of the complexes of eRF3 PAM2-N (yellow) and PAM2-C (magenta) bound to the MLLE domain (grey) from PABPC1. eRF3 Phe76 shifts position and binds to either MLLE helix α2/α3 in the PAM2-N complex or helix α3/α5 in the PAM2-C complex. Leu69 or Phe85 then occupies the vacated hydrophobic binding site. (C) Stick representation of overlaid structures of eRF3 PAM2-N (yellow) and PAM2-C (magenta) bound to the MLLE domain from PABPC1 (green).